 | |
| Names | |
|---|---|
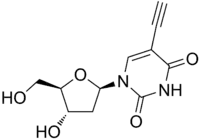
| IUPAC name
2′-Deoxy-5-ethynyluridine | |
| Systematic IUPAC name
5-Ethynyl-1-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)oxolan-2-yl]pyrimidine-2,4(1H,3H)-dione | |
| Other names
5-Ethynyl-2′-deoxyuridine | |
| Identifiers | |
3D model (JSmol) |
|
| Abbreviations | EdU |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.230.902 |
| MeSH | 5-ethynyl-2'-deoxyuridine |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C11H12N2O5 | |
| Molar mass | 252.226 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
5-Ethynyl-2′-deoxyuridine (EdU) is a thymidine analogue which is incorporated into the DNA of dividing cells. EdU is used to assay DNA synthesis in cell culture and detect cells in embryonic, neonatal and adult animals which have undergone DNA synthesis.[1] Whilst at high doses it can be cytotoxic, this molecule is now widely used to track proliferating cells in multiple biological systems.
EdU-labelling allows cells to be isolated without denaturing DNA, allowing researchers to determine the transcriptional profile of cells.[2] This approach has been used to assess transcription in neuronal cells[3] and tissues[4] that have recently divided either in vitro or in vivo.
Detection
EdU is labeled and detected with an azide molecule (most commonly fluorescent azides) through Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) click chemistry.[1][5] Unlike the commonly used bromodeoxyuridine (BrdU), EdU detection requires no heat or acid treatment.
Toxicity
EdU incorporated into DNA induces DNA damage through the formation of interstrand crosslinks.[6] These are detected by the cell during DNA replication, which is reflected by phosphorylation of histone H2AX, arrest in the cell cycle progression, and apoptosis.[7]
References
- 1 2 Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G, Mackay-Sim A (February 2009). "EdU, a new thymidine analogue for labelling proliferating cells in the nervous system". Journal of Neuroscience Methods. 177 (1): 122–130. doi:10.1016/j.jneumeth.2008.10.006. hdl:10072/25744. PMID 18996411. S2CID 21427874.
- ↑ Endaya B, Cavanagh B, Alowaidi F, Walker T, de Pennington N, Ng JM, et al. (January 2016). "Isolating dividing neural and brain tumour cells for gene expression profiling". Journal of Neuroscience Methods. 257: 121–133. doi:10.1016/j.jneumeth.2015.09.020. PMID 26432933. S2CID 44969376.
- ↑ Endaya BB, Lam PY, Meedeniya AC, Neuzil J (January 2016). "Transcriptional profiling of dividing tumor cells detects intratumor heterogeneity linked to cell proliferation in a brain tumor model". Molecular Oncology. 10 (1): 126–137. doi:10.1016/j.molonc.2015.09.001. PMC 5528932. PMID 26388584.
- ↑ Ng HX, Lee EP, Cavanagh BL, Britto JM, Tan SS (September 2017). "A method for isolating cortical interneurons sharing the same birthdays for gene expression studies". Experimental Neurology. 295: 36–45. doi:10.1016/j.expneurol.2017.05.006. PMID 28511841. S2CID 3377296.
- ↑ Cavanagh BL, Walker T, Norazit A, Meedeniya AC (September 2011). "Thymidine analogues for tracking DNA synthesis". Molecules. 16 (9): 7980–7993. doi:10.3390/molecules16097980. PMC 6264245. PMID 21921870.
- ↑ Ligasová A, Strunin D, Friedecký D, Adam T, Koberna K (2015-02-11). "A fatal combination: a thymidylate synthase inhibitor with DNA damaging activity". PLOS ONE. 10 (2): e0117459. doi:10.1371/journal.pone.0117459. PMC 4324964. PMID 25671308.
- ↑ Zhao H, Halicka HD, Li J, Biela E, Berniak K, Dobrucki J, Darzynkiewicz Z (November 2013). "DNA damage signaling, impairment of cell cycle progression, and apoptosis triggered by 5-ethynyl-2'-deoxyuridine incorporated into DNA". Cytometry. Part A. 83 (11): 979–988. doi:10.1002/cyto.a.22396. PMC 3846616. PMID 24115313.
External links
 Media related to EdU at Wikimedia Commons
Media related to EdU at Wikimedia Commons