Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid[1] binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation,[2][3] compared to other chromatographic methods.
Principle
Affinity chromatography has the advantage of specific binding interactions between the analyte of interest (normally dissolved in the mobile phase), and a binding partner or ligand (immobilized on the stationary phase). In a typical affinity chromatography experiment, the ligand is attached to a solid, insoluble matrix—usually a polymer such as agarose or polyacrylamide—chemically modified to introduce reactive functional groups with which the ligand can react, forming stable covalent bonds.[4] The stationary phase is first loaded into a column to which the mobile phase is introduced. Molecules that bind to the ligand will remain associated with the stationary phase. A wash buffer is then applied to remove non-target biomolecules by disrupting their weaker interactions with the stationary phase, while the biomolecules of interest will remain bound. Target biomolecules may then be removed by applying a so-called elution buffer, which disrupts interactions between the bound target biomolecules and the ligand. The target molecule is thus recovered in the eluting solution.[5]
Affinity chromatography does not require the molecular weight, charge, hydrophobicity, or other physical properties of the analyte of interest to be known, although knowledge of its binding properties is useful in the design of a separation protocol.[5] Types of binding interactions commonly exploited in affinity chromatography procedures are summarized in the table below.
| Sr. no | Types of ligand | Target molecule |
|---|---|---|
| 1 | Substrate analogue | Enzymes |
| 2 | Antibody | Antigen |
| 3 | Lectin | Polysaccharide |
| 4 | Nucleic acid | Complementary base sequence |
| 5 | Hormone | Receptor |
| 6 | Avidin | Biotin/Biotin-conjugated molecule |
| 7 | Calmodulin | Calmodulin binding partner |
| 8 | Glutathione | GST fusion protein |
| 9 | Protein A or Protein G | Immunoglobulins |
| 10 | Nickel-NTA | polyhistidine fusion protein |
Batch and column setups

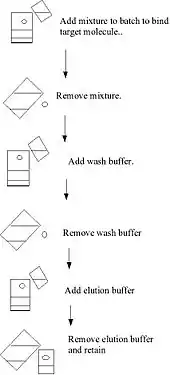
Binding to the solid phase may be achieved by column chromatography whereby the solid medium is packed onto a column, the initial mixture run through the column to allow settling, a wash buffer run through the column and the elution buffer subsequently applied to the column and collected. These steps are usually done at ambient pressure. Alternatively, binding may be achieved using a batch treatment, for example, by adding the initial mixture to the solid phase in a vessel, mixing, separating the solid phase, removing the liquid phase, washing, re-centrifuging, adding the elution buffer, re-centrifuging and removing the elute.
Sometimes a hybrid method is employed such that the binding is done by the batch method, but the solid phase with the target molecule bound is packed onto a column and washing and elution are done on the column.
The ligands used in affinity chromatography are obtained from both organic and inorganic sources. Examples of biological sources are serum proteins, lectins and antibodies. Inorganic sources are moronic acid, metal chelates and triazine dyes.[7]
A third method, expanded bed absorption, which combines the advantages of the two methods mentioned above, has also been developed. The solid phase particles are placed in a column where liquid phase is pumped in from the bottom and exits at the top. The gravity of the particles ensure that the solid phase does not exit the column with the liquid phase.
Affinity columns can be eluted by changing salt concentrations, pH, pI, charge and ionic strength directly or through a gradient to resolve the particles of interest.
More recently, setups employing more than one column in series have been developed. The advantage compared to single column setups is that the resin material can be fully loaded since non-binding product is directly passed on to a consecutive column with fresh column material. These chromatographic processes are known as periodic counter-current chromatography (PCC). The resin costs per amount of produced product can thus be drastically reduced. Since one column can always be eluted and regenerated while the other column is loaded, already two columns are sufficient to make full use of the advantages.[8] Additional columns can give additional flexibility for elution and regeneration times, at the cost of additional equipment and resin costs.
Specific uses
Affinity chromatography can be used in a number of applications, including nucleic acid purification, protein purification[9] from cell free extracts, and purification from blood.
By using affinity chromatography, one can separate proteins that bind to a certain fragment from proteins that do not bind that specific fragment.[10] Because this technique of purification relies on the biological properties of the protein needed, it is a useful technique and proteins can be purified many folds in one step.[11]
Various affinity media
Many different affinity media exist for a variety of possible uses.[12][9][13] Briefly, they are (generalized) activated/functionalized that work as a functional spacer, support matrix, and eliminates handling of toxic reagents.
Amino acid media is used with a variety of serum proteins, proteins, peptides, and enzymes, as well as rRNA and dsDNA. Avidin biotin media is used in the purification process of biotin/avidin and their derivatives.
Carbohydrate bonding is most often used with glycoproteins or any other carbohydrate-containing substance; carbohydrate is used with lectins, glycoproteins, or any other carbohydrate metabolite protein. Dye ligand media is nonspecific but mimics biological substrates and proteins. Glutathione is useful for separation of GST tagged recombinant proteins. Heparin is a generalized affinity ligand, and it is most useful for separation of plasma coagulation proteins, along with nucleic acid enzymes and lipases
Hydrophobic interaction media are most commonly used to target free carboxyl groups and proteins.
Immunoaffinity media (detailed below) utilizes antigens' and antibodies' high specificity to separate; immobilized metal affinity chromatography is detailed further below and uses interactions between metal ions and proteins (usually specially tagged) to separate; nucleotide/coenzyme that works to separate dehydrogenases, kinases, and transaminases.
Nucleic acids function to trap mRNA, DNA, rRNA, and other nucleic acids/oligonucleotides. Protein A/G method is used to purify immunoglobulins.
Speciality media are designed for a specific class or type of protein/co enzyme; this type of media will only work to separate a specific protein or coenzyme.
Immunoaffinity
Another use for the procedure is the affinity purification of antibodies from blood serum. If the serum is known to contain antibodies against a specific antigen (for example if the serum comes from an organism immunized against the antigen concerned) then it can be used for the affinity purification of that antigen. This is also known as Immunoaffinity Chromatography. For example, if an organism is immunised against a GST-fusion protein it will produce antibodies against the fusion-protein, and possibly antibodies against the GST tag as well. The protein can then be covalently coupled to a solid support such as agarose and used as an affinity ligand in purifications of antibody from immune serum.
For thoroughness, the GST protein and the GST-fusion protein can each be coupled separately. The serum is initially allowed to bind to the GST affinity matrix. This will remove antibodies against the GST part of the fusion protein. The serum is then separated from the solid support and allowed to bind to the GST-fusion protein matrix. This allows any antibodies that recognize the antigen to be captured on the solid support. Elution of the antibodies of interest is most often achieved using a low pH buffer such as glycine pH 2.8. The eluate is collected into a neutral tris or phosphate buffer, to neutralize the low pH elution buffer and halt any degradation of the antibody's activity. This is a nice example as affinity purification is used to purify the initial GST-fusion protein, to remove the undesirable anti-GST antibodies from the serum and to purify the target antibody.
Monoclonal antibodies can also be selected to bind proteins with great specificity, where protein is released under fairly gentle conditions. This can become of use for further research in the future.[14]
A simplified strategy is often employed to purify antibodies generated against peptide antigens. When the peptide antigens are produced synthetically, a terminal cysteine residue is added at either the N- or C-terminus of the peptide. This cysteine residue contains a sulfhydryl functional group which allows the peptide to be easily conjugated to a carrier protein (e.g. Keyhole limpet hemocyanin (KLH)). The same cysteine-containing peptide is also immobilized onto an agarose resin through the cysteine residue and is then used to purify the antibody.
Most monoclonal antibodies have been purified using affinity chromatography based on immunoglobulin-specific Protein A or Protein G, derived from bacteria.[15]
Immunoaffinity chromatography with monoclonal antibodies immobilized on monolithic column has been successfully used to capture extracellular vesicles (e.g., exosomes and exomeres) from human blood plasma by targeting tetraspanins and integrins found on the surface of the EVs.[16][17]
Immunoaffinity chromatography is also the basis for immunochromatographic test (ICT) strips, which provide a rapid means of diagnosis in patient care. Using ICT, a technician can make a determination at a patient's bedside, without the need for a laboratory.[18] ICT detection is highly specific to the microbe causing an infection.[19]
Immobilized metal ion affinity chromatography
Immobilized metal ion affinity chromatography (IMAC) is based on the specific coordinate covalent bond of amino acids, particularly histidine, to metals. This technique works by allowing proteins with an affinity for metal ions to be retained in a column containing immobilized metal ions, such as cobalt, nickel, or copper for the purification of histidine-containing proteins or peptides, iron, zinc or gallium for the purification of phosphorylated proteins or peptides. Many naturally occurring proteins do not have an affinity for metal ions, therefore recombinant DNA technology can be used to introduce such a protein tag into the relevant gene. Methods used to elute the protein of interest include changing the pH, or adding a competitive molecule, such as imidazole.[20][21]

Recombinant proteins
Possibly the most common use of affinity chromatography is for the purification of recombinant proteins. Proteins with a known affinity are protein tagged in order to aid their purification. The protein may have been genetically modified so as to allow it to be selected for affinity binding; this is known as a fusion protein. Protein tags include hexahistidine (His), glutathione-S-transferase (GST), maltose binding protein (MBP), and the Colicin E7 variant CL7 tag. Histidine tags have an affinity for nickel, cobalt, zinc, copper and iron ions which have been immobilized by forming coordinate covalent bonds with a chelator incorporated in the stationary phase. For elution, an excess amount of a compound able to act as a metal ion ligand, such as imidazole, is used. GST has an affinity for glutathione which is commercially available immobilized as glutathione agarose. During elution, excess glutathione is used to displace the tagged protein. CL7 has an affinity and specificity for Immunity Protein 7 (Im7) which is commercially available immobilized as Im7 agarose resin. For elution, an active and site-specific protease is applied to the Im7 resin to release the tag-free protein.[22]
Lectins
Lectin affinity chromatography is a form of affinity chromatography where lectins are used to separate components within the sample. Lectins, such as concanavalin A are proteins which can bind specific alpha-D-mannose and alpha-D-glucose carbohydrate molecules. Some common carbohydrate molecules that is used in lectin affinity chromatography are Con A-Sepharose and WGA-agarose.[23] Another example of a lectin is wheat germ agglutinin which binds D-N-acetyl-glucosamine.[24] The most common application is to separate glycoproteins from non-glycosylated proteins, or one glycoform from another glycoform.[25] Although there are various ways to perform lectin affinity chromatography, the goal is extract a sugar ligand of the desired protein.[23]
Specialty
Another use for affinity chromatography is the purification of specific proteins using a gel matrix that is unique to a specific protein. For example, the purification of E. coli β-galactosidase is accomplished by affinity chromatography using p-aminobenyl-1-thio-β-D-galactopyranosyl agarose as the affinity matrix. p-aminobenyl-1-thio-β-D-galactopyranosyl agarose is used as the affinity matrix because it contains a galactopyranosyl group, which serves as a good substrate analog for E. coli β-Galactosidase. This property allows the enzyme to bind to the stationary phase of the affinity matrix and β-Galactosidase is eluted by adding increasing concentrations of salt to the column.[26]
Alkaline phosphatase
Alkaline phosphatase from E. coli can be purified using a DEAE-Cellulose matrix. A. phosphatase has a slight negative charge, allowing it to weakly bind to the positively charged amine groups in the matrix. The enzyme can then be eluted out by adding buffer with higher salt concentrations.[27]
Boronate affinity chromatography
Boronate affinity chromatography consists of using boronic acid or boronates to elute and quantify amounts of glycoproteins. Clinical adaptations have applied this type of chromatography for use in determining long term assessment of diabetic patients through analysis of their glycated hemoglobin.[24]
Serum albumin purification
Affinity purification of albumin and macroglobulin contamination is helpful in removing excess albumin and α2-macroglobulin contamination, when performing mass spectrometry. In affinity purification of serum albumin, the stationary used for collecting or attracting serum proteins can be Cibacron Blue-Sepharose. Then the serum proteins can be eluted from the adsorbent with a buffer containing thiocyanate (SCN−).[28]
Weak affinity chromatography
Weak affinity chromatography[29] (WAC) is an affinity chromatography technique for affinity screening in drug development.[30][31] WAC is an affinity-based liquid chromatographic technique that separates chemical compounds based on their different weak affinities to an immobilized target. The higher affinity a compound has towards the target, the longer it remains in the separation unit, and this will be expressed as a longer retention time. The affinity measure and ranking of affinity can be achieved by processing the obtained retention times of analyzed compounds. Affinity chromatography is part of a larger suite of techniques used in chemoproteomics based drug target identification.
The WAC technology is demonstrated against a number of different protein targets – proteases, kinases, chaperones and protein–protein interaction (PPI) targets. WAC has been shown to be more effective than established methods for fragment based screening.[31]
History
Affinity chromatography was conceived and first developed by Pedro Cuatrecasas and Meir Wilchek.[32][33]
References
- ↑ Aizpurua-Olaizola, Oier; Sastre Torano, Javier; Pukin, Aliaksei; Fu, Ou; Boons, Geert Jan; de Jong, Gerhardus J.; Pieters, Roland J. (January 2018). "Affinity capillary electrophoresis for the assessment of binding affinity of carbohydrate-based cholera toxin inhibitors". Electrophoresis. 39 (2): 344–347. doi:10.1002/elps.201700207. PMID 28905402. S2CID 33657660.
- ↑ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2009). Fundamental Laboratory Approaches for Biochemistry and Biotechnology (2nd ed.). Wiley. p. 133. ISBN 9780470087664.
- ↑ ""Introduction to Affinity Chromatography"". bio-rad.com. Bio-Rad. 14 September 2020. Retrieved 14 September 2020.
- ↑ Zachariou, Michael, ed. (2008). Affinity Chromatography: Methods and Protocols (2nd ed.). Totowa, N.J.: Humana Press. pp. 1–2. ISBN 9781588296597.
- 1 2 Bonner, Philip L.R. (2007). Protein Purification (2nd ed.). Totowa, N.J.: Taylor & Francis Group. ISBN 9780415385114.
- ↑ Kumar, Pranav (2018). Biophysics and Molecular Biology. New Delhi: Pathfinder Publication. p. 11. ISBN 978-93-80473-15-4.
- ↑ Fanali, Salvatore; Haddad, Paul R.; Poole, Colin F.; Schoenmakers, Peter; Lloyd, David, eds. (2013). Liquid Chromatography: Applications. Handbooks in Separation Science. Saint Louis: Elsevier. p. 3. ISBN 9780124158061.
- ↑ Baur, Daniel; Angarita, Monica; Müller-Späth, Thomas; Steinebach, Fabian; Morbidelli, Massimo (2016). "Comparison of batch and continuous multi-column protein A capture processes by optimal design". Biotechnology Journal. 11 (7): 920–931. doi:10.1002/biot.201500481. hdl:11311/1013726. PMID 26992151. S2CID 205492204.
- 1 2 "Cube Biotech". Cube Biotech. Retrieved 11 September 2019.
- ↑ Ahern, Kevin (12 February 2015). Biochemistry Free & Easy. DaVinci Press; 3rd Edition. p. 822.
- ↑ Grisham, Charles M. (1 January 2013). Biochemistry. Brooks/Cole, Cengage Learning. ISBN 978-1133106296. OCLC 777722371.
- ↑ Mahmoudi Gomari, Mohammad; Saraygord-Afshari, Neda; Farsimadan, Marziye; Rostami, Neda; Aghamiri, Shahin; Farajollahi, Mohammad M. (1 December 2020). "Opportunities and challenges of the tag-assisted protein purification techniques: Applications in the pharmaceutical industry". Biotechnology Advances. 45: 107653. doi:10.1016/j.biotechadv.2020.107653. ISSN 0734-9750. PMID 33157154. S2CID 226276355.
- ↑ "Affinity Chromatography".
- ↑ Thompson, Nancy E.; Foley, Katherine M.; Stalder, Elizabeth S.; Burgess, Richard R. (2009). "Chapter 28 Identification, Production, and Use of Polyol-Responsive Monoclonal Antibodies for Immunoaffinity Chromatography". Guide to Protein Purification, 2nd Edition. Methods in Enzymology. Vol. 463. pp. 475–494. doi:10.1016/s0076-6879(09)63028-7. ISBN 9780123745361. PMID 19892188.
- ↑ Uhlén M (2008). "Affinity as a tool in life science". BioTechniques. 44 (5): 649–54. doi:10.2144/000112803. PMID 18474040.
- ↑ Multia E, Tear CJ, Palviainen M, et al. (December 2019). "Fast isolation of highly specific population of platelet-derived extracellular vesicles from blood plasma by affinity monolithic column, immobilized with anti-human CD61 antibody". Analytica Chimica Acta. 1091: 160–168. doi:10.1016/j.aca.2019.09.022. hdl:10138/321264. PMID 31679569. S2CID 203147714.
- ↑ Multia E, Liangsupree T, Jussila M, et al. (September 2020). "Automated On-Line Isolation and Fractionation System for Nanosized Biomacromolecules from Human Plasma". Analytical Chemistry. 92 (19): 13058–13065. doi:10.1021/acs.analchem.0c01986. PMC 7586295. PMID 32893620.
- ↑ Luppa, Peter (2018). Point-of-care testing: principles and clinical applications. Berlin, Germany: Springer. pp. 71–72. ISBN 9783662544976.
- ↑ J. D. Muller; C. R. Wilks; K. J. O'Riley; R. J. Condron; R. Bull; A. Mateczun (2004). "Specificity of an immunochromatographic test for anthrax". Australian Veterinary Journal. 82 (4): 220–222. doi:10.1111/j.1751-0813.2004.tb12682.x. PMID 15149073.
- ↑ Singh, Naveen K.; DSouza, Roy N.; Bibi, Noor S.; Fernández-Lahore, Marcelo (2015). "Direct Capture of His6-Tagged Proteins Using Megaporous Cryogels Developed for Metal-Ion Affinity Chromatography". In Reichelt, S. (ed.). Affinity Chromatography. Methods in Molecular Biology. Vol. 1286. New York: Humana Press. pp. 201–212. doi:10.1007/978-1-4939-2447-9_16. ISBN 978-1-4939-2447-9. PMID 25749956.
- ↑ Gaberc-Porekar, Vladka K.; Menart, Viktor (2001). "Perspectives of immobilized-metal affinity chromatography". J Biochem Biophys Methods. 49 (1–3): 335–360. doi:10.1016/S0165-022X(01)00207-X. PMID 11694288.
- ↑ Vassylyeva MN, Klyuyev S, Vassylyev AD, Wesson H, Zhang Z, Renfrow MB, Wang H, Higgins NP, Chow LT, Vassylyev DG. Efficient, ultra-high-affinity chromatography in a one-step purification of complex proteins. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5138-E5147. doi: 10.1073/pnas.1704872114. Epub 2017 Jun 12. PMID: 28607052; PMCID: PMC5495267.
- 1 2 Freeze, H. H. (May 2001). "Lectin affinity chromatography". Current Protocols in Protein Science. Vol. Chapter 9. pp. 9.1.1–9.1.9. doi:10.1002/0471140864.ps0901s00. ISBN 978-0471140863. ISSN 1934-3663. PMID 18429210. S2CID 3197260.
- 1 2 Hage, David (May 1999). "Affinity Chromatography: A Review of Clinical Applications" (PDF). Clinical Chemistry. 45 (5): 593–615. doi:10.1093/clinchem/45.5.593. PMID 10222345.
- ↑ "GE Healthcare Life Sciences, Immobilized lectin". Archived from the original on 3 March 2012. Retrieved 29 November 2010.
- ↑ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2006). Fundamental Laboratory Approaches for Biochemistry and Biotechnology (2nd ed.). Wiley. p. 153.
- ↑ Ninfa, Alexander J.; Ballou, David P.; Benore, Marilee (2010). Fundamental laboratory approaches for biochemistry and biotechnology (2nd ed.). Hoboken, N.J.: John Wiley. p. 240. ISBN 9780470087664. OCLC 420027217.
- ↑ Naval, Javier; Calvo, Miguel; Lampreave, Fermin; Piñeiro, Andrés (1 January 1983). "Affinity chromatography of serum albumin: An illustrative laboratory experiment on biomolecular interactions". Biochemical Education. 11 (1): 5–8. doi:10.1016/0307-4412(83)90004-3. ISSN 1879-1468.
- ↑ Zopf, D.; S. Ohlson (1990). "Weak-affinity chromatography". Nature. 346 (6279): 87–88. Bibcode:1990Natur.346...87Z. doi:10.1038/346087a0. ISSN 0028-0836. S2CID 4306269.
- ↑ Duong-Thi, M. D.; Meiby, E.; Bergström, M.; Fex, T.; Isaksson, R.; Ohlson, S. (2011). "Weak affinity chromatography as a new approach for fragment screening in drug discovery". Analytical Biochemistry. 414 (1): 138–146. doi:10.1016/j.ab.2011.02.022. PMID 21352794.
- 1 2 Meiby, E.; Simmonite, H.; Le Strat, L.; Davis, B.; Matassova, N.; Moore, J. D.; Mrosek, M.; Murray, J.; Hubbard, R. E.; Ohlson, S. (2013). "Fragment Screening by Weak Affinity Chromatography: Comparison with Established Techniques for Screening against HSP90". Analytical Chemistry. 85 (14): 6756–6766. doi:10.1021/ac400715t. PMID 23806099.
- ↑ "Meir Wilchek - Wolf Foundation". Wolf Foundation. 9 December 2018. Retrieved 17 March 2021.
Affinity chromatography is a novel technique which was conceived by Cuatrecasas and Wilchek
- ↑ P Cuatrecasas; M Wilchek; C B Anfinsen (October 1968). "Selective enzyme purification by affinity chromatography". Proceedings of the National Academy of Sciences of the United States of America. 61 (2): 636–643. Bibcode:1968PNAS...61..636C. doi:10.1073/pnas.61.2.636. PMC 225207. PMID 4971842.
External links
- "Affinity Chromatography Principle, Procedure And Advance Detailed Note – 2020".
- "What is affinity chromatography"
| Library resources about Affinity chromatography |