Glutamate receptor-interacting protein (GRIP) refers to either a family of proteins that bind to the glutamate receptor or specifically to the GRIP1 protein within this family. Proteins in the glutamate receptor-interacting protein (GRIP) family have been shown to interact with GluR2, a common subunit in the AMPA receptor.[1] This subunit also interacts with other proteins such as protein interacting with C-kinase1 (PICK1) and N-ethylmaleimide-sensitive fusion protein (NSF). Studies have begun to elucidate its function; however, much is still to be learned about these proteins.
Discovery and history of GRIP 1

The discovery of the Glutamate Receptor Interacting Protein (GRIP-1) came as a result of the observation that Glutamate Receptors, such as the NMDA receptor, cluster at synapses.[2] Shortly after this observation, researchers identified a region on the C-terminal region of NMDA receptors called the tSXV motif that has the ability to bind to the PDZ domain of the PSD-95 protein.[3]
Research on NMDA receptor localization paved the way for research on non-NMDA receptors such as AMPA receptors. Similar to NMDA receptors, it was discovered that AMPA receptors localize in the synaptic terminal of neurons in the central nervous system.[4] By using GFP (green fluorescent protein) antibodies that correspond to the GRIP protein, researchers were able to use fluorescence to determine the location of GRIP in hippocampal neurons. Another GFP antibody was then used to label the GluR2 subunit of AMPA receptors.[4] By using immunocytochemistry and comparing the location of GRIP and AMPA receptors it was determined that GRIP and AMPA receptors experience colocalization in hippocampal neurons.[4] These findings confirmed the initial hypothesis that the GRIP protein plays an important role in binding AMPA receptors to excitatory synapses.
The structure of GRIP contains seven PDZ domains and binds to the C-terminus of the GluR2 subunit of AMPA receptors.[4] Although the number of PDZ domains is different for the proteins PSD-95 and GRIP, the PDZ domain is a common structural motif in proteins that help mediate protein-protein interactions.[5] The AMPA receptor amino acid sequence that the GRIP protein binds to is ESVKI. The conserved serine amino acid in the C- terminus of both AMPA and NMDA receptors suggests that it plays an important role in facilitating the interaction for GRIP and PSD-95.[6]
Role of GRIP in AMPAR cycling
AMPA receptors are constantly being transported between the cell membrane and intracellular space and it was originally thought that GRIP may be responsible for the clustering of AMPA receptors at the excitatory synapse.[1] Although it is still unclear the exact role of GRIP in this trafficking, It appears that PICK1 is more directly responsible for the clustering of AMPA receptors at the surface and that GRIP is involved in the stabilization of AMPA receptors intracellularly.[7] One study showed that when the interaction between GluR2 and GRIP is disrupted, there are no changes in the surface expression of AMPA receptors or the constitutive internalization of AMPA receptors.[8] There is, however, a reduced amount of receptors that remain internalized when receptor cycling is modified by application of AMPA-1. The ratio returns to normal when constitutive recycling is allowed to happen, suggesting that the stabilization of intracellular receptors is critical only under AMPA-induced internalization.[8]

In later studies, two proteins, GRIP-1 (often reduced to GRIP) and ABP-L (also named GRIP-2), were found to be expressed by two separate genes and their respective contributions to AMPA receptor cycling have since been well studied. Each of these proteins have different isoforms due to differential RNA splicing.[9][10] The isoforms of GRIP-1 are named GRIP-1a and GRIP-1b while those of ABP-L are distinguished as ABP-L and pABP-L. The apparent difference in both cases is that one isoform (GRIP1b and pABP-L respectively) is capable of being conjugated with Palmitic acid, an action called Palmitoylation.
Whereas GRIP initially was thought to be involved in the stabilization of AMPA receptors either at the cell surface or intracellularly when internalization was triggered by AMPA stimulation, it now appears that the GRIP-1 isoforms are involved differentially with the stabilization of AMPA receptors after being internalized due to NMDA stimulation.[11] GRIP-1a has been shown to reduce the expected intracellular levels of AMPA receptors after NMDA stimulation. Conversely, GRIP-1b increases intracellular levels of AMPA receptors under the same conditions.
ABP-L, like GRIP-1b, associates with intracellular stores of AMPA receptors. pABP-L, however, associates with AMPA receptors as the surface membrane.[12] It has not yet been shown under what conditions these interactions are significant in the cycling of AMPAR.
Role of GRIP1 in Fraser syndrome
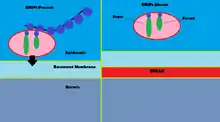
Mutations to GRIP1 play a role in less than 10% of confirmed cases of the group of congenital defects known as Fraser syndrome.[13] Using immunofluorescence, it has been shown that GRIP1 is found in several kinds of embryonic tissues, including the GI tract, ureter buds, skin and oral and nasal cavities.[14] GRIP1 is also essential for proper function and structure of the dermo-epidermal junction.[15] In mouse models, knocking out GRIP1 protein leads to several deformities that begin in embryo. These deformities include subepidermal hemorrhagic blistering, renal agenesis, syndactylism, polydactylism and cryptopthalmos.[14] One study has shown that complete knock-out of GRIP1 leads to the absence of kidneys.[14] Another study shows blistering of embryonic tissue that GRIP1 is expressed in by day 12 of embryonic life in mice.[15]
The mechanism of GRIP1 in Fraser syndrome is found in the interaction GRIP1 has with the proteins Fras1 and Frem2.[16] Fras1 and Frem2 are extracellular membrane proteins necessary for proper basement membrane function as well as morphogenesis.[16] GRIP1 plays a vital role in localizing Fras1 to the basal surface of epidermal cells as well as localizing Frem2.[16] Knocking out the GRIP1 protein or mutating it leads to poor expression of Fras1 and Frem2.[16] GRIP1 specifically binds with Fras1 through a PDZ motif located on Fras1.[16] Frem2 also has a PDZ domain, although the interaction between GRIP1 and Frem2 is unclear.[16] In one case of Fraser syndrome, GRIP1 lacked PDZ domains 6 and 7. Only the first four PDZ domains of the seven PDZ domains GRIP1 has are required for binding with Fras1, indicating additional mechanisms and proteins GRIP1 interacts with that could lead to Fraser syndrome when mutated.[17] Other mutations in GRIP1 that lead to Fraser syndrome include nonsense mutations, frameshift mutations, splice site mutations, a genome deletion and a deletion of exon 18 of the GRIP1 gene.[13]
Role of GRIP1 in neuron morphology and cargo transport
Neuron morphology, development, and maintenance are dependent on the expression of GRIP1 in the cell.[18] It is vitally important in initial development as knock out experiments in murine models result in skin blisters and embryonic lethality.[15] In developed murine models, disabling mutations like transfecton or dominant negatives in GRIP1 can cause up to 75% loss in “primary, secondary, and higher-order” dendrites in developing neurons.[19] Disabling GRIP1 in live healthy neurons in a dish will cause a 20% reduction in the thickest part of the neuron and up to 70% reduction in the branches.[18]
Defects in neuron morphology due to GRIP1 malfunction can be reserved. One way is to overexpress GRIP1. This leads to increased, but not complete recovery of branching.[18] Another protein, EphB2, which interacts with GRIP1, can be mutated such that a 70-90% recovery of branching is possible. However, overexpression of the wild type leads to a decrease in neuron count.[18]
Motor proteins such as Kinesin (KIF5) are bound to adapter molecules like GRIP1 to move cargo from the Golgi to the extremities of a neuron cell. GRIP1 and KIF5 are very commonly found together due to a good binding affinity (Kd range from 10-20nM[20]). As for how cargo gets to the right place, there has been a hypothesis called the “smart motor”.[21] It is currently thought that the “smart motor” recognizes the difference between axonal (coated with KLC protein) and dendritic (coated with KHC) proteins.[21] The destination is chosen accordingly. Unfortunately, details about the intermediate transporting steps are unknown. However, at the destination the binding of protein 14-3-3 disrupts the interaction between KIF5 and GRIP1.[19] This releases the cargo.
See also
References
- 1 2 Dong, H; O'Brien, R; Fung, E; Lanahan, A; Worley, P; Hunganir, R (March 1997). "GRIP: A synaptic PDZ domain-containing protein that interacts with AMPA receptors". Nature. 386 (6622): 279–84. Bibcode:1997Natur.386..279D. doi:10.1038/386279a0. PMID 9069286. S2CID 4361791.
- ↑ Kornau, Hans-Christian; Schenker, Leslie T.; Kennedy, Mary B.; Seeburg, Peter H. (1995). "Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95". Science. 269 (5231): 1737–1740. Bibcode:1995Sci...269.1737K. doi:10.1126/science.7569905. PMID 7569905.
- ↑ Ehlers, M. D.; Mammen, A. L.; Lau, L.; Huganir, R. L. (1996). "Synaptic targeting of glutamate receptors". Current Opinion in Cell Biology. 8 (4): 484–489. doi:10.1016/S0955-0674(96)80024-X. PMID 8791455.
- 1 2 3 4 Dong, H; O'Brien, R; Fung, E; Lanahan, A; Worley, P; Hunganir, R (1997). "GRIP: A synaptic PDZ domain-containing protein that interacts with AMPA receptors". Nature. 386 (6622): 279–84. Bibcode:1997Natur.386..279D. doi:10.1038/386279a0. PMID 9069286. S2CID 4361791.
- ↑ Lee, HJ; Zheng, JJ (2010). "PDZ domains and their binding partners: structure, specificity, and modification". Cell Commun Signal. 8: 8. doi:10.1186/1478-811X-8-8. PMC 2891790. PMID 20509869.
- ↑ Ehlers, M.; Mammen, A.; Lau, L.; Huganir, R. (1998). "Synaptic targeting of glutamate receptors". Current Opinion in Cell Biology. 8 (4): 484–489. doi:10.1016/S0955-0674(96)80024-X. PMID 8791455.
- ↑ Xia, J; Zhang, X; Staudinger, J; Huganir, R (January 1999). "Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1". Neuron. 22 (1): 179–87. doi:10.1016/s0896-6273(00)80689-3. PMID 10027300.
- 1 2 Braithwaite, S; Xia, H; Malenka, R (2002). "Differential roles for NSF and GRIP/ABP in AMPA receptor cycling". Proceedings of the National Academy of Sciences. 99 (10): 7096–101. Bibcode:2002PNAS...99.7096B. doi:10.1073/pnas.102156099. PMC 124534. PMID 12011465.
- ↑ Dong, H; Zhang, P; Song, I; Petralia, R; Liao, D; Hunganir, R (15 August 1999). "Characterization of the glutamate receptor-interacting proteins GRIP1 and GRIP2". Journal of Neuroscience. 19 (16): 6930–41. doi:10.1523/JNEUROSCI.19-16-06930.1999. PMC 6782851. PMID 10436050.
- ↑ Wyszynski, M; Valtschanoff, J; Naisbitt, S; Dunah, A; Kim, E; Standaert, D; et al. (1 August 1999). "Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo". Journal of Neuroscience. 19 (15): 6528–37. doi:10.1523/JNEUROSCI.19-15-06528.1999. PMC 6782830. PMID 10414981.
- ↑ Hanley, L; Henley, J (2010). "Differential roles of GRIP1a and GRIP1b in AMPA receptor trafficking". Neuroscience Letters. 485 (3): 167–72. doi:10.1016/j.neulet.2010.09.003. PMC 3310156. PMID 20837103.
- ↑ DeSouza, S; Fu, J; States, B; Ziff, E (2002). "Differential palmitoylation directs the AMPA receptor-binding protein ABP to spines or to intracellular clusters". Journal of Neuroscience. 22 (9): 3493–503. doi:10.1523/JNEUROSCI.22-09-03493.2002. PMC 6758378. PMID 11978826.
- 1 2 Schanze, D (2014). "Fraser syndrome due to mutations in GRIP1--clinical phenotype in two families and expansion of the mutation spectrum". American Journal of Medical Genetics. 164 (3): 837–840. doi:10.1002/ajmg.a.36343. PMID 24357607. S2CID 11034897.
- 1 2 3 Takamiya, K.; Kostourou, V.; Adams, S.; Jadeja, S.; Chalepakis, G.; Scambler, P. J.; Adams, R. H. (2004). "A direct functional link between the multi-PDZ domain protein GRIP1 and the fraser syndrome protein Fras1". Nature Genetics. 36 (2): 172–7. doi:10.1038/ng1292. PMID 14730302.
- 1 2 3 Bladt, F (2002). "Epidermolysis bullosa and embryonic lethality in mice lacking the multi-PDZ domain protein GRIP1". Proceedings of the National Academy of Sciences of the United States of America. 99 (10): 6816–6821. Bibcode:2002PNAS...99.6816B. doi:10.1073/pnas.092130099. PMC 124486. PMID 11983858.
- 1 2 3 4 5 6 Kiyozumi, D (2006). "Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes fraser syndrome-like defects". Proceedings of the National Academy of Sciences of the United States of America. 103 (32): 11981–11986. Bibcode:2006PNAS..10311981K. doi:10.1073/pnas.0601011103. PMC 1567684. PMID 16880404.
- ↑ Vogel, M., Zon, P., Brueton, L., Gijzen, M., Tuil, M., Cox, P., ... Haelst, M. (2012). Mutations in GRIP1 cause Fraser syndrome. Journal of Medical Genetics, 303-306.
- 1 2 3 4 Hoogenraad, Casper (Jun 19, 2005). "GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking". Nature Neuroscience. 8 (7): 906–916. doi:10.1038/nn1487. PMID 15965473. S2CID 23686585.
- 1 2 Geiger, Julia (Feb 24, 2014). "The GRIP1/14-3-3 Pathway Coordinates Cargo Trafficking and Dendrite Development". Developmental Cell. 28 (4): 381–393. doi:10.1016/j.devcel.2014.01.018. PMID 24576423.
- ↑ Skoufias, Dimitrios (Jan 14, 1994). "The Carboxyl-terminal Domain of Kinesin Heavy Chain is important for membrane binding". Journal of Biological Chemistry. 269 (2): 1477–1485. doi:10.1016/S0021-9258(17)42281-2. PMID 8288613.
- 1 2 Setou, Mitsutoshi (May 2, 2002). "Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites". Letters to Nature. 417 (6884): 83–87. Bibcode:2002Natur.417...83S. doi:10.1038/nature743. PMID 11986669. S2CID 4400494.