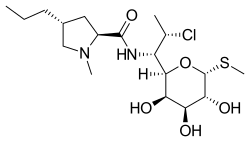
Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin.[1]
Structure
Lincosamides consist of a pyrrolidine ring linked to a pyranose moiety (methylthio-lincosamide) via an amide bond.[2][3] Hydrolysis of lincosamides, specifically lincomycin, splits the molecule into its building blocks of the sugar and proline moieties. Both of these derivatives can conversely be recombined into the drug itself or a derivative.[4]
Synthesis
Biosynthesis of lincosamides occurs through a biphasic pathway, in which propylproline and methylthiolincosamide are independently synthesized immediately before condensation of the two precursor molecules. Condensation of the propylproline carboxyl group with the methylthiolincosamide amine group via an amide bond forms N-demethyllincomycin. N-Demethyllincomycin is subsequently methylated via S-adenosyl methionine to produce lincomycin A.[5][6]
Lincomycin is naturally produced by bacteria species, namely Streptomyces lincolnensis, S. roseolus, and S. caelestis.[7] Clindamycin is derived via (7S)-chloro-substitution of the (7R)-hydroxyl group of lincomycin.[8] Lincomycin is primarily isolated from fermentations of Streptomyces lincolnensis, while clindamycin is prepared semi-synthetically.[9] While several hundred synthetic and semi-synthetic derivatives of lincosamides have been prepared, only lincomycin A and clindamycin are used in clinical practice due to issues with toxicity and low biological activity in other lincosamide antibiotics.[9]

Mechanism of action
Lincosamides prevent bacterial replication in a bacteriostatic mechanism by interfering with the synthesis of proteins.
In a mechanism similar to macrolides and streptogramin B, lincosamides bind close to the peptidyl transferase center on the 23S portion of the 50S subunit of bacterial ribosomes. Under the influence of high resolution X-ray, structures of clindamycin and ribosomal subunits from bacterium have previously revealed exclusive binding to the 23S segment of the peptidyl transferase cavity.[10] Binding is mediated by the mycarose sugar moiety which has partially overlapping substrates with peptidyl transferase. By extending to the peptidyl transferase center, lincosamides cause the premature dissociation of peptidyl-tRNA's containing two, three or four amino acid residues. In this case, peptides will grow to a certain point until steric hindrance inhibits peptidyl transferase activity.[11] Lincosamides do not interfere with protein synthesis in human cells (or those of other eukaryotes) due to structural differences between prokaryotic and eukaryotic ribosomes. Lincosamides are used against Gram-positive bacteria since they are unable to pass through the porins of Gram-negative bacteria.

Resistance
Ribosomal methylation
Soon after the emergence of clinical lincosamide use in 1953, strains of resistant staphylococci were isolated in several countries including France, Japan and the United States.[13] Resistant strains were characterized by expression of methyltransferases which dimethylate residues within the 23S subunit of ribosomal RNA, preventing binding of macrolides, lincosamides and streptogramins B. The gene family responsible for encoding of these methyltransferases is referred to as the "erm" family, or erythromycin ribosome methylase family of genes.[14] Nearly 40 erm genes have been reported to date, which are transferred primarily through plasmids and transposons.[15]
Target mutation
Several strains of bacteria which are highly resistant to macrolide treatment have been isolated and found to possess mutations at the transferase binding pocket in the 23S ribosomal subunit. Macrolide-resistant Streptococcus pneumoniae isolated from hospital patients in Eastern Europe and North America were found to contain mutations in either 23S or other ribosomal protein genes.[16]
Antibiotic efflux
Gram-negative bacteria harbor genes encoding for molecular pumps which can contribute to resistance of hydrophobic compounds like macrolides and lincosamides.[14] Out of the many families of multidrug resistance pumps, lincosamides are most commonly shunted through pumps belonging to the resistance-nodulation-cell division superfamily.[17] Staphylococci express efflux pumps with specificity for 14 and 15 member ring macrolides and streptogramin B, but not lincosamide molecules.[18]
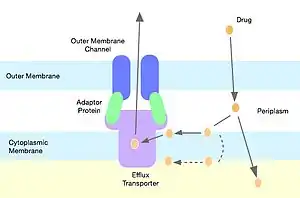
Drug modification
Clinical isolates of S. aureus harboring genes which encode for lincosamide nucleotransferases have been reported. Genes lnuA and lnuB confer resistance to lincomycin, but not clindamycin. These genes, however, limit the bacteriostatic activity of clindamycin.[15] This type of resistance is rare in S. aureus, but has been reported to be more prevalent in other bacteria strains.[19]
Pharmacokinetics
Approximately 90% of orally administered lincosamides are absorbed, with slight variance depending on which drug is given. Plasma concentrations via this route peak within 2–4 hours. Intramuscular administration of lincosamides results in strong absorption, with peak plasma levels being reached in 1–2 hours. Around 90% of clindamycin is bound to plasma proteins, and is generally more stable and rapidly absorbed than lincomycin.[20]
Lincosamides have a broad distribution in several tissues, excluding cerebrospinal fluid. When administered intramuscularly to rats, lincomycin was found to accumulate in highest concentrations in the kidneys when compared to other tissues, while clindamycin was found in highest concentrations within the lungs.[21] Clindamycin accumulates in macrophages and other white blood cells, which can result in concentrations 50 times higher than plasma levels.[22]
Clinical use
Lincosamides are often used clinically as an alternative antibiotic for patients who are allergic to penicillin. Of the lincosamides, clindamycin is most commonly used within the clinic due to its higher bioavailability, higher oral absorption and efficacy within the target organism spectrum.[23] Lincosamides are generally the first-choice use antibiotic class in veterinary microbiology, most commonly used to combat skin infections.[7]
Potential clinical uses for lincosamide antibiotics in humans are numerous. They are efficacious in the treatment of dental infections, abdominal infections, abscesses, pelvic inflammatory disease and anaerobic infections. Clindamycin alone has been shown to be efficacious in the treatment of acne,[24] toxic shock syndrome[25] and malaria,[26] and to decrease the risk of premature births in women with bacterial vaginosis.[27] Lincosamide antibiotics may also be useful in the treatment of methicillin-resistant S. aureus.[28]
Toxicity and interactions

While there have been no reports of severe organ toxicity from lincosamide treatment, gastrointestinal disturbances have been associated with their administration. Pseudomembranous enterocolitis resulting from clindamycin-induced disruption of gastrointestinal flora can be a lethal adverse event observed in several species when used in the veterinary clinic, particularly in horses. At extremely high doses of clindamycin, skeletal muscle paralysis has been demonstrated in several species. Lincosamides can interact with anesthetic agents to produce neuromuscular effects.[29]
Other adverse reactions include diarrhea, nausea, vomiting, abdominal pain and rash. Topical administration of clindamycin may induce contact dermatitis, dryness, burning, itching, scaliness and peeling of the skin.[30]
Lincosamide brand name formulations
- Clindamycin - Cleocin, Cleocin Pediatric, ClindaMax Vaginal, Dalacin
- Lincomycin - Lincocin
- Pirlimycin
History
The first lincosamide compound discovered was lincomycin, isolated from Streptomyces lincolnensis in a soil sample from Lincoln, Nebraska (hence the bacterial name).[2]
Further reading
- Van Bambeke F. Mechanisms of action. In Armstrong D, Cohen J. Infectious diseases. Mosby, London, 1999, pp7/1.1-7/1.14
References
- ↑ Sonia Ilaria Maffioli (2014). "A Chemist's Survey of Different Antibiotic Classes". In Claudio O. Gualerzi; Letizia Brandi; Attilio Fabbretti; Cynthia L. Pon (eds.). Antibiotics: Targets, Mechanisms and Resistance. Wiley-VCH. ISBN 9783527659685.
- 1 2 Rezanka, Tomas; Spizek, Jaroslav; Sigler, Karel (2007-04-01). "Medicinal Use of Lincosamides and Microbial Resistance to Them". Anti-Infective Agents in Medicinal Chemistry. 6 (2): 133–144. doi:10.2174/187152107780361670. ISSN 1871-5214.
- ↑ Pubchem. "Lincosamides". pubchem.ncbi.nlm.nih.gov. Retrieved 2018-09-19.
- ↑ Le Goffic, Francois (1 January 1985). "Structure activity relationships in lincosamide and streptogramin antibiotics". Journal of Antimicrobial Chemotherapy. 16 (suppl A): 13–21. doi:10.1093/jac/16.suppl_a.13. PMID 3932299.
- ↑ Rezanka, Tomas; Spizek, Jaroslav; Sigler, Karel (2007). "Medicinal Use of Lincosamides and Microbial Resistance to Them". Anti-Infective Agents in Medicinal Chemistry. 6 (2): 133–144. doi:10.2174/187152107780361670. Retrieved 2018-10-07.
- ↑ "ucsdchem257 / Lincomycin". ucsdchem257.pbworks.com. Retrieved 2018-11-10.
- 1 2 Spížek, Jaroslav; Řezanka, Tomáš (2017-06-01). "Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications". Biochemical Pharmacology. 133: 20–28. doi:10.1016/j.bcp.2016.12.001. ISSN 0006-2952. PMID 27940264. S2CID 21224168.
- ↑ Birkenmeyer, Robert D.; Kagan, Fred. (July 1970). "Lincomycin. XI. Synthesis and structure of clindamycin, a potent antibacterial agent". Journal of Medicinal Chemistry. 13 (4): 616–619. doi:10.1021/jm00298a007. ISSN 0022-2623. PMID 4916317.
- 1 2 Spížek, J.; Řezanka, T. (2004-02-05). "Lincomycin, clindamycin and their applications". Applied Microbiology and Biotechnology. 64 (4): 455–464. doi:10.1007/s00253-003-1545-7. ISSN 0175-7598. PMID 14762701. S2CID 7870760.
- ↑ Schlünzen, Frank; Zarivach, Raz; Harms, Jörg; Bashan, Anat; Tocilj, Ante; Albrecht, Renate; Yonath, Ada; Franceschi, François (October 2001). "Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria". Nature. 413 (6858): 814–821. Bibcode:2001Natur.413..814S. doi:10.1038/35101544. ISSN 0028-0836. PMID 11677599. S2CID 205022511.
- ↑ The Mechanism of Action of Macrolides, Lincosamides and Streptogramin B Reveals the Nascent Peptide Exit Path in the Ribosome Martin Lovmar and Måns Ehrenberg
- ↑ Tenson, Tanel; Lovmar, Martin; Ehrenberg, Måns (2003-07-25). "The Mechanism of Action of Macrolides, Lincosamides and Streptogramin B Reveals the Nascent Peptide Exit Path in the Ribosome". Journal of Molecular Biology. 330 (5): 1005–1014. doi:10.1016/S0022-2836(03)00662-4. ISSN 0022-2836. PMID 12860123.
- ↑ Weisblum, B (March 1995). "Erythromycin resistance by ribosome modification". Antimicrobial Agents and Chemotherapy. 39 (3): 577–585. doi:10.1128/aac.39.3.577. ISSN 0066-4804. PMC 162587. PMID 7793855.
- 1 2 Leclercq, Roland (February 2002). "Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications". Clinical Infectious Diseases. 34 (4): 482–492. doi:10.1086/324626. ISSN 1058-4838. PMID 11797175. S2CID 14714107.
- 1 2 Roberts, Marilyn C.; Sutcliffe, Joyce; Courvalin, Patrice; Jensen, Lars Bogo; Rood, Julian; Seppala, Helena (1999-12-01). "Nomenclature for Macrolide and Macrolide-Lincosamide-Streptogramin B Resistance Determinants". Antimicrobial Agents and Chemotherapy. 43 (12): 2823–2830. doi:10.1128/AAC.43.12.2823. ISSN 0066-4804. PMC 89572. PMID 10582867.
- ↑ Tait-Kamradt, A.; Davies, T.; Appelbaum, P. C.; Depardieu, F.; Courvalin, P.; Petitpas, J.; Wondrack, L.; Walker, A.; Jacobs, M. R. (2000-12-01). "Two New Mechanisms of Macrolide Resistance in Clinical Strains of Streptococcus pneumoniae from Eastern Europe and North America". Antimicrobial Agents and Chemotherapy. 44 (12): 3395–3401. doi:10.1128/AAC.44.12.3395-3401.2000. ISSN 0066-4804. PMC 90211. PMID 11083646.
- ↑ Yılmaz, Çiğdem; Özcengiz, Gülay (2017-06-01). "Antibiotics: Pharmacokinetics, toxicity, resistance and multidrug efflux pumps". Biochemical Pharmacology. 133: 43–62. doi:10.1016/j.bcp.2016.10.005. ISSN 0006-2952. PMID 27765485. S2CID 25336534.
- ↑ Sutcliffe, J; Tait-Kamradt, A; Wondrack, L (August 1996). "Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system". Antimicrobial Agents and Chemotherapy. 40 (8): 1817–1824. doi:10.1128/aac.40.8.1817. ISSN 0066-4804. PMC 163423. PMID 8843287.
- ↑ Bozdogan, Bülent; Berrezouga, Latifa; Kuo, Ming-Shang; Yurek, David A.; Farley, Kathleen A.; Stockman, Brian J.; Leclercq, Roland (1999-04-01). "A New Resistance Gene, linB, Conferring Resistance to Lincosamides by Nucleotidylation in Enterococcus faecium HM1025". Antimicrobial Agents and Chemotherapy. 43 (4): 925–929. doi:10.1128/AAC.43.4.925. ISSN 0066-4804. PMC 89227. PMID 10103201.
- ↑ "Lincosamides - Pharmacology - Merck Veterinary Manual". Merck Veterinary Manual. Retrieved 2018-11-10.
- ↑ Osono, T.; Umezawa, H. (July 1985). "Pharmacokinetics of macrolides, lincosamides and streptogramins". The Journal of Antimicrobial Chemotherapy. 16 Suppl A: 151–166. doi:10.1093/jac/16.suppl_a.151. ISSN 0305-7453. PMID 3932301.
- ↑ Johnson, J. D.; Hand, W. L.; Francis, J. B.; King-Thompson, N.; Corwin, R. W. (March 1980). "Antibiotic uptake by alveolar macrophages". The Journal of Laboratory and Clinical Medicine. 95 (3): 429–439. ISSN 0022-2143. PMID 7354244.
- ↑ Greenwood, D.; Irving, W.L. (2012-01-01). "Antimicrobial agents". Medical Microbiology: 54–68. doi:10.1016/B978-0-7020-4089-4.00020-2. ISBN 9780702040894.
- ↑ Leyden, J. J.; Berger, R. S.; Dunlap, F. E.; Ellis, C. N.; Connolly, M. A.; Levy, S. F. (2001). "Comparison of the efficacy and safety of a combination topical gel formulation of benzoyl peroxide and clindamycin with benzoyl peroxide, clindamycin and vehicle gel in the treatments of acne vulgaris". American Journal of Clinical Dermatology. 2 (1): 33–39. doi:10.2165/00128071-200102010-00006. ISSN 1175-0561. PMID 11702619. S2CID 22486823.
- ↑ Annane, Djillali; Clair, Bernard; Salomon, Jérôme (August 2004). "Managing toxic shock syndrome with antibiotics". Expert Opinion on Pharmacotherapy. 5 (8): 1701–1710. doi:10.1517/14656566.5.8.1701. ISSN 1744-7666. PMID 15264985. S2CID 24494787.
- ↑ Lell, Bertrand; Kremsner, Peter G. (August 2002). "Clindamycin as an Antimalarial Drug: Review of Clinical Trials". Antimicrobial Agents and Chemotherapy. 46 (8): 2315–2320. doi:10.1128/AAC.46.8.2315-2320.2002. ISSN 0066-4804. PMC 127356. PMID 12121898.
- ↑ Lamont, Ronnie F. (March 2005). "Can antibiotics prevent preterm birth--the pro and con debate". BJOG: An International Journal of Obstetrics and Gynaecology. 112 (Suppl 1): 67–73. doi:10.1111/j.1471-0528.2005.00589.x. ISSN 1470-0328. PMID 15715599. S2CID 25572794.
- ↑ "Sitemap". Archived from the original on 2011-10-08.
- ↑ "Lincosamides - Pharmacology - Merck Veterinary Manual". Merck Veterinary Manual. Retrieved 2018-11-23.
- ↑ de Groot, Mark C H; van Puijenbroek, Eugène P (October 2007). "Clindamycin and taste disorders". British Journal of Clinical Pharmacology. 64 (4): 542–545. doi:10.1111/j.1365-2125.2007.02908.x. ISSN 0306-5251. PMC 2048568. PMID 17635503.
