Outer sphere refers to an electron transfer (ET) event that occurs between chemical species that remain separate and intact before, during, and after the ET event.[1] In contrast, for inner sphere electron transfer the participating redox sites undergoing ET become connected by a chemical bridge. Because the ET in outer sphere electron transfer occurs between two non-connected species, the electron is forced to move through space from one redox center to the other.
Marcus theory
The main theory that describes the rates of outer sphere electron transfer was developed by Rudolph A. Marcus in the 1950s. A major aspect of Marcus theory is the dependence of the electron transfer rate on the thermodynamic driving force (difference in the redox potentials of the electron-exchanging sites). For most reactions, the rates increase with increased driving force. A second aspect is that the rate of outer sphere electron-transfer depends inversely on the "reorganizational energy." Reorganization energy describes the changes in bond lengths and angles that are required for the oxidant and reductant to switch their oxidation states. This energy is assessed by measurements of the self-exchange rates (see below).
Outer sphere electron transfer is the most common type of electron transfer, especially in biochemistry, where redox centers are separated by several (up to about 11) angstroms by intervening protein. In biochemistry, there are two main types of outer sphere ET: ET between two biological molecules or fixed distance electron transfer, in which the electron transfers within a single biomolecule (e.g., intraprotein).[2]
Examples
Self-exchange
Outer sphere electron transfer can occur between chemical species that are identical except for their oxidation state.[3] This process is termed self-exchange. An example is the degenerate reaction between the tetrahedral ions permanganate and manganate:
- [MnO4]− + [Mn*O4]2− → [MnO4]2− + [Mn*O4]−
For octahedral metal complexes, the rate constant for self-exchange reactions correlates with changes in the population of the eg orbitals, the population of which most strongly affects the length of metal-ligand bonds:
- For the [Co(bipy)3]+/[Co(bipy)3]2+ pair, self exchange proceeds at 109 M−1s−1. In this case, the electron configuration changes from Co(I): (t2g)6(eg)2 to Co(II): (t2g)5(eg)2.
- For the [Co(bipy)3]2+/[Co(bipy)3]3+ pair, self exchange proceeds at 18 M−1s−1. In this case, the electron configuration changes from Co(II): (t2g)5(eg)2 to Co(III): (t2g)6(eg)0.
Iron-sulfur proteins
Outer sphere ET is the basis of the biological function of the iron-sulfur proteins. The Fe centers are typically further coordinated by cysteinyl ligands. The [Fe4S4] electron-transfer proteins ([Fe4S4] ferredoxins) may be further subdivided into low-potential (bacterial-type) and high-potential (HiPIP) ferredoxins. Low- and high-potential ferredoxins are related by the following redox scheme:
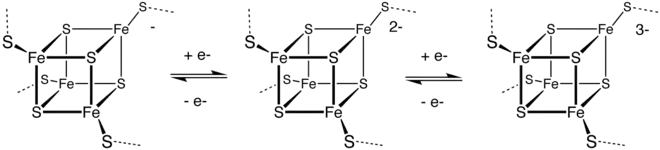
Because of the small structural differences between the individual redox states, ET is rapid between these clusters.
See also
References
- ↑ Article: outer-sphere electron transfer, from the IUPAC Gold book]
- ↑ S. J. Lippard, J. M. Berg “Principles of Bioinorganic Chemistry” University Science Books: Mill Valley, CA; 1994 ISBN 0-935702-73-3
- ↑ R. G. Wilkins Kinetics and Mechanism of Reactions of Transition Metal Complexes, 2nd Edition, VCH, Weinheim, 1991. ISBN 1-56081-125-0