| B3 DNA binding domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
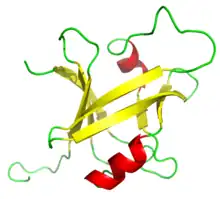 B3 DNA binding domain of RAV1 | |||||||||||
| Identifiers | |||||||||||
| Symbol | B3_domain | ||||||||||
| Pfam | PF02362 | ||||||||||
| InterPro | IPR003340 | ||||||||||
| PROSITE | PS50863 | ||||||||||
| SCOP2 | 1wid / SCOPe / SUPFAM | ||||||||||
| CDD | cd10017 | ||||||||||
| |||||||||||
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors (≥40 species) (Pfam PF02362) combined with other domains (InterPro: IPR003340). It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold (SCOP 117343); it interacts with the major groove of DNA.[1]
B3 families
In Arabidopsis thaliana, there are three main families of transcription factors that contain B3 domain:[2]
- ARF (Auxin Response Factors)
- ABI3 (ABscisic acid Insensitive3)
- RAV (Related to ABI3/VP1)
| protein | ARF1-B3 | ABI3-B3 | RAV1-B3 |
|---|---|---|---|
| B3 structure derived by | molecular model[1] | molecular model[1] | NMR[1] |
| B3 recognition sequence | TGTCTC[3][4] | CATGCA[5][6] | CACCTG[7] |
PDB: 1WID[1] and PDB: 1YEL[8] are only known NMR solution phase structures of the B3 DNA Binding Domain.
Related proteins
The N-terminal domain of restriction endonuclease EcoRII; the C-terminal domain of restriction endonuclease BfiI possess a similar DNA-binding pseudobarrel protein fold.[9][10]
See also
- Restriction endonuclease EcoRII
- Auxin
- Abscisic acid
References
- 1 2 3 4 5 Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Hayami N, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S (2004). "Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1". Plant Cell. 16 (12): 3448–59. doi:10.1105/tpc.104.026112. PMC 535885. PMID 15548737.
- ↑ Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000). "Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes". Science. 290 (5499): 2105–10. Bibcode:2000Sci...290.2105R. doi:10.1126/science.290.5499.2105. PMID 11118137.
- ↑ Ulmasov T, Hagen G, Guilfoyle TJ (1997). "ARF1, a transcription factor that binds to auxin response elements". Science. 276 (5320): 1865–8. doi:10.1126/science.276.5320.1865. PMID 9188533.
- ↑ Tiwari SB, Hagen G, Guilfoyle TJ (2003). "The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription". Plant Cell. 15 (2): 533–43. doi:10.1105/tpc.008417. PMC 141219. PMID 12566590.
- ↑ Suzuki M, Kao CY, McCarty DR (1997). "The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity". Plant Cell. 9 (5): 799–807. doi:10.1105/tpc.9.5.799. PMC 156957. PMID 9165754.
- ↑ Ezcurra I, Wycliffe P, Nehlin L, Ellerström M, Rask L (2000). "Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box". Plant J. 24 (1): 57–66. doi:10.1046/j.1365-313x.2000.00857.x. PMID 11029704.
- ↑ Kagaya Y, Ohmiya K, Hattori T (1999). "RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants". Nucleic Acids Res. 27 (2): 470–8. doi:10.1093/nar/27.2.470. PMC 148202. PMID 9862967.
- ↑ Waltner, J.K.; Peterson, F.C.; Lytle, B.L.; Volkman, B.F. (2005). "Structure of the B3 domain from Arabidopsis thaliana protein At1g16640". Protein Sci. 14 (9): 2478–83. doi:10.1110/ps.051606305. PMC 2253459. PMID 16081658.
- ↑ Zhou XE, Wang Y, Reuter M, Mücke M, Krüger DH, Meehan EJ, Chen L (2004). "Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold". J. Mol. Biol. 335 (1): 307–19. doi:10.1016/j.jmb.2003.10.030. PMID 14659759.
- ↑ Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V (2005). "Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease" (PDF). Proc. Natl. Acad. Sci. U.S.A. 102 (44): 15797–802. Bibcode:2005PNAS..10215797G. doi:10.1073/pnas.0507949102. PMC 1266039. PMID 16247004.
External links
- DBD database of predicted transcription factors Archived 2008-12-04 at the Wayback MachineKummerfeld SK, Teichmann SA (2006). "DBD: a transcription factor prediction database". Nucleic Acids Res. 34 (Database issue): D74–81. doi:10.1093/nar/gkj131. PMC 1347493. PMID 16381970. Uses a curated set of DNA-binding domains to predict transcription factors in all completely sequenced genomes
- Classification in the "Transcription factors" table according to the Transfac database.
- Database of Arabidopsis Transcription Factors
- B3 Archived 2019-10-17 at the Wayback Machine, RAV, and ARF Archived 2019-10-17 at the Wayback Machine family at PlantTFDB:Plant Transcription Factor Database