 | |
| Names | |
|---|---|
| Preferred IUPAC name
(6aR,11aS,11bR)-10-Acetyl-11-hydroxy-7,7-dimethyl-2,6,6a,7,11a,11b-hexahydro-9H-pyrrolo[1′,2′:2,3]isoindolo[4,5,6-cd]indol-9-one | |
| Identifiers | |
3D model (JSmol) |
|
| 707309 | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.162.058 |
| EC Number |
|
| KEGG | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C20H20N2O3 | |
| Molar mass | 336.391 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Cyclopiazonic acid (α-CPA), a mycotoxin and a fungal neurotoxin, is made by the molds Aspergillus and Penicillium.[1][2][3] It is an indole-tetramic acid that serves as a toxin due to its ability to inhibit calcium-dependent ATPases found in the endoplasmic and sarcoplasmic reticulum.[4] This inhibition disrupts the muscle contraction-relaxation cycle and the calcium gradient that is maintained for proper cellular activity in cells.[2]
Cyclopiazonic acid is known to contaminate multiple foods because the molds that produce them are able to grow on different agricultural products, including but not limited to grains, corn, peanuts, and cheese.[2][5] Due to this contamination, α-CPA can be harmful to both humans and farm animals that were exposed to contaminated animal feeds. However, α-CPA needs to be introduced in very high concentrations to produce mycotoxicosis in animals. Due to this, α-CPA is not a potent acute toxin.[2]
Chemically, CPA is related to ergoline alkaloids. CPA was originally isolated from Penicillium cyclopium and subsequently from other fungi including Penicillium griseofulvum, Penicillium camemberti, Penicillium commune, Aspergillus flavus, and Aspergillus versicolor. CPA only appears to be toxic in high concentrations. Ingestion of CPA causes anorexia, dehydration, weight loss, immobility, and signs of spasm when near death. CPA can be found in molds, corns, peanuts, and other fermented products, such as cheese and sausages.[6] Biologically, CPA is a specific inhibitor of SERCA ATPase in intracellular Ca2+ storage sites.[7] CPA inhibits SERCA ATPase by keeping it in one specific conformation, thus, preventing it from forming another.[8] CPA also binds to SERCA ATPase at the same site as another inhibitor, thapsigargin (TG). In this way, CPA lowers the ability of SERCA ATPase to bind an ATP molecule.[9]
Toxicity
Cases of α-CPA mycotoxicosis in humans are rare. However, the occurrence of α-CPA in foods consumed by humans suggests that the toxin is indeed ingested by humans, though at concentrations low enough to be of no serious health concern.[10] Even if its toxicity in humans is rare, large doses of α-CPA have been seen to adversely affect animals such as mice, rats, chickens, pigs, dogs, and rabbits.[5] Cyclopiazonic acid's toxicity mirrors that of antipsychotic drugs when taken up these animals.[5] This mycotoxin has been extensively studied in mice to discern its toxic properties. The severity of toxicity is dose-dependent, and exposure to α-CPA has led to hypokinesia, hypothermia, catalepsy, tremors, irregular respiration, ptosis, weight loss, and eventual death in mice.[5] The adverse health effects of α-CPA studied in mice are similar to those found in other animals.
Biosynthesis
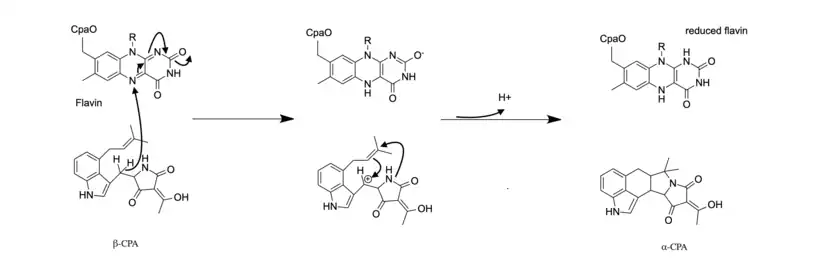
Three enzymes are utilized in the biosynthesis of α-CPA: the polypeptide CpaS, dimethylallyltransferase (CpaD), and flavoprotein oxidocyclase (CpaO).[3] CpaS is the first enzyme in the biosynthetic pathway and is a hybrid polyketide synthase- nonribosomal peptide synthetase (PKS-NRPS). It uses the precursors acetyl-CoA, malonyl-CoA, and tryptophan to produce cyclo-acetoaceytl-L-tryptophan (cAATrp).[3] The intermediate cAATrp is then prenylated with dimethylallyl pyrophosphate (DMAPP) by the enzyme CpaD to form the intermediate β-CPA. CpaD has high substrate specificity and will not catalyze prenylation in the presence of DMAPP's isomer isopentyl pyrophosphate (IPP) or the derivatives of cAATrp.[2] The third enzyme, CpaO, then acts on β-CPA through a redox mechanism that allows for intramolecular cyclization to form α-CPA.[3]

Mechanism of Action of CpaS
CpaS is made of several domains that belong either to the PKS portion or the NRPS portion of the 431 kDa protein.[2][3] The PKS portion is made up of three catalytically important domains and three additional tailoring domains that are common to polyketide synthases but not used in the biosynthesis of α-CPA. The catalytically important acyl carrier protein domain (ACP), acyl transferase domain (AT), and ketosynthase domain (KS) work together to form acetoacetyl-CoA from the precursors acetyl-CoA and malonyl-CoA.[2] The acetoacetyl-CoA is then acted on by the NRPS portion of CpaS. The NRPS portion, like the PKS portion, contains many catalytically active domains. The adenylation domain (A) acts first to activate the amino acid tryptophan and subsequently transfer it to the peptidyl carrier protein (PCP) domain (T).[2] Following this, the condensation domain (C) catalyzes an amide bond formation between the acetoacetyl moiety attached to the ACP and tryptophan attached to the PCP.[2] The releasing domain (R) catalyzes a Dieckmann condensation to both cyclize and release the cAATrp product.[2][3]
Formation of β-CPA
The second enzyme, CpaD, converts the cAATrp produced by CpaS to β-CPA. CpaD, also known as cycloacetoacetyltyptophanyl dimethylallyl transferase, places DMAPP at the tryptophan indole ring, specifically at position C-4.[2] CpaD then catalyzes selective prenylation at position C-4 through a Friedel-Craft alkylation, producing β-CPA.[2] It is important to note here that the biosynthesis of α-CPA is dependent on other pathways, specifically the mevalonate pathway, which serves to form DMAPP.[2]
Formation of α-CPA
The final enzyme in the biosynthetic pathway, CpaO, converts β-CPA to α-CPA. CpaO is a FAD-dependent oxidoreductase. FAD oxidizes β-CPA in a two-electron process, subsequently allowing for ring closure and formation of α-CPA.[2] To regenerate the oxidized FAD cofactor used by CpaO, the reduced FAD reacts with molecular oxygen to produce hydrogen peroxide.
References
- ↑ Holzapfel CW (March 1968). "The isolation and structure of cyclopiazonic acid, a toxic metabolite of Penicillium cyclopium Westling". Tetrahedron. 24 (5): 2101–19. doi:10.1016/0040-4020(68)88113-X. PMID 5636916.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Chang PK, Ehrlich KC, Fujii I (December 2009). "Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae". Toxins. 1 (2): 74–99. doi:10.3390/toxins1020074. PMC 3202784. PMID 22069533.
- 1 2 3 4 5 6 Liu X, Walsh CT (September 2009). "Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan". Biochemistry. 48 (36): 8746–57. doi:10.1021/bi901123r. PMC 2752376. PMID 19663400.
- ↑ Seidler NW, Jona I, Vegh M, Martonosi A (October 1989). "Cyclopiazonic acid is a specific inhibitor of the Ca2+-ATPase of sarcoplasmic reticulum". The Journal of Biological Chemistry. 264 (30): 17816–23. doi:10.1016/S0021-9258(19)84646-X. PMID 2530215.
- 1 2 3 4 Nishie K, Cole RJ, Dorner JW (September 1985). "Toxicity and neuropharmacology of cyclopiazonic acid". Food and Chemical Toxicology. 23 (9): 831–9. doi:10.1016/0278-6915(85)90284-4. PMID 4043883.
- ↑ Bullerman LB (2003). "MYCOTOXINS | Classifications". Encyclopedia of Food Sciences and Nutrition. pp. 4080–4089. doi:10.1016/B0-12-227055-X/00821-X. ISBN 978-0-12-227055-0.
- ↑ Sosa MJ, Córdoba JJ, Díaz C, Rodríguez M, Bermúdez E, Asensio MA, Núñez F (June 2002). "Production of cyclopiazonic acid by Penicillium commune isolated from dry-cured ham on a meat extract-based substrate". Journal of Food Protection. 65 (6): 988–92. doi:10.4315/0362-028X-65.6.988. PMID 12092733.
- ↑ Soler F, Plenge-Tellechea F, Fortea I, Fernandez-Belda F (March 1998). "Cyclopiazonic acid effect on Ca2+-dependent conformational states of the sarcoplasmic reticulum ATPase. Implication for the enzyme turnover". Biochemistry. 37 (12): 4266–74. doi:10.1021/bi971455c. PMID 9521749.
- ↑ Ma H, Zhong L, Inesi G, Fortea I, Soler F, Fernandez-Belda F (November 1999). "Overlapping effects of S3 stalk segment mutations on the affinity of Ca2+-ATPase (SERCA) for thapsigargin and cyclopiazonic acid". Biochemistry. 38 (47): 15522–7. doi:10.1021/bi991523q. PMID 10569935.
- ↑ Voss KA (May 1990). "In Vivo and In Vitro Toxicity of Cyclopiazonic Acid (CPA)". In Llewellyn GC, O'Rear CE (eds.). Biodeterioration Research: Mycotoxins, Biotoxins, Wood Decay, Air Quality, Cultural Properties, General Biodeterioration, and Degradation. Biodeterioration Research. Boston, MA: Springer US. pp. 67–84. doi:10.1007/978-1-4757-9453-3_5. ISBN 978-1-4757-9453-3.