| DHX8 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | DHX8, DDX8, HRH1, PRP22, PRPF22, DEAH-box helicase 8, Dhr2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 600396 MGI: 1306823 HomoloGene: 3628 GeneCards: DHX8 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
DEAH-box helicase 8, is a protein that in humans is encoded by the DHX8 gene. This protein is member of the DEAH box polypeptide family. The main characteristic of this group is their conserved motif DEAH (Asp- Glu- Ala- His).[5] A wide range of RNA helicases belongs to this family. Specifically, DHX8 acts as an ATP-dependent RNA helicase involved in splicing and the regulation of the releasing of spliced mRNAs from spliceosomes out of the nucleus.[6] Published studies have shown the consequences of DHX8 mutations, some of them are critical for biological processes such as hematopoiesis and are related to some diseases.[7][8]
Structure
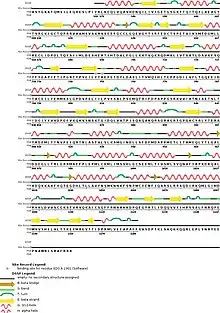
Protein DHX8 is part of a protein complex called spliceosome, which is in charge of pre-mRNA splicing. The spliceosome has eight major functional states, each with distinct composition and structure; five of the eight states have been structurally characterized.[10]
DHX8 have different domains: a S1 RNA binding domain (DEAD/DEAH box), an helicase conserved C-terminal domain, helicase associated domain (HA2), and an oligonucleotide/oligosaccharide-binding (OB)-fold, each joined by intrinsically disordered regions.[11]
There are some regions of the protein which are very important for its activity, like R620 and the hook-loop and hook-turn regions. Also, DHX8Δ547 is the catalytically active core of the protein DHX8. It is made of two RecA domains and the C-terminal WH, ratchet-like and OB-fold domains and the N-terminal region.[9]
The total weight of the DHX8 structure is 156580.13 Da.
Secondary structure:
- 36% helical: 26 helices and 246 residues
- 16% beta sheet: 29 strands and 110 residues.[9]
Function
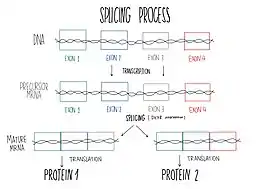
DHX8 is localized in the cellular nucleus and stimulated upon RNA presence. This protein is a component of the spliceosome, so it takes part in pre-mRNA splicing.[9] Splicing is the process of joining exons from primary transcripts of messenger RNA and the elimination of intron sequences, by means of a spliceosomal mechanism, so that the mRNA produced is the one without introns, consisting exclusively of the joined exons.[12] Splicing finishes with the spliceosomal complex disassembly and the ATP-dependent liberation of the resulting mature RNAs to the outer of the nucleus.[13] Spliceosome requires conformational changes to be able to catalyze splicing reactions and the later mature mRNA releasing to the outer of the nucleus. One of the ATP-dependent helicase needed for these conformational changes is DHX8. Furthermore, DHX8 plays a key role in the releasing, facilitating the nuclear export of spliced mRNA. Protein characterization has shown that DHX8 has a binding preference for adenine-rich RNA. This binding is followed by ATP hydrolysis and thus, ADP release.[9]
Mechanism
DHX8 has multiple molecular functions like ATP-binding by, selective and non-covalent interactions with the coenzyme and enzyme regulator adenosine 5’ triphosphate.[14] Also identical protein binding (creates a similar type of interactions as described above but with other proteins), RNA binding and RNA helicase activity, based on catalysis of the reaction that unwinds an RNA helix:[15]
ATP + H2O = ADP + phosphate[16]
Tissues distribution

DHX8 protein has its main focuses of expression in the brain, especially in the cerebellum where it can be found mostly in Purkinje cells. Other important are the prostate and the gallbladder, which are zones where the polypetide is found also highly expressed.
DHX8 protein is not expressed in all tissues and organs significantly, some clear examples are the bone marrow or the soft tissue (Peripheral nerve), where we do not have enough quantity of the protein to be representative.[17]
Purification and cloning
Assays in enzymology for the biochemical characterization of proteins need high concentrations of the protein of interest and its protocols should be efficient, simple and cost-efficient to ensure a successful purification. One example of a purification approach for DHX8 is via a protein tag called GST-His grafted onto DHX8 protein is used.
The N-terminal Glutathione Sepharose TAG (GST) and C-termianl His-tag, also known as GST-His is a 29 kDa tag which allows small-scale affinity purification for recombinant proteins. This method is based on two different tags flanking the two extremes of the protein. However, it might influence physiological properties of the protein and thus, empirical testing is required for each case.[18]
DHX8 constructs are generated by PCR cloning using restriction enzymes. To generate His6GST-DHX8Δ54 (hexahistidine-GST), the coding sequence for residues A548 to R1220 is inserted into a version of "pFastBac", a specific vector kit of Thermo Fisher Scientific,[19] modified to encode an N-terminal His6GST-tag followed by an HRV 3C protease (recombinant restriction-grade protease) cleavage site. HRV 3C is a highly purified recombinant 6XHis-fusion protein, that recognizes the same cleavage site as the native enzyme. Both, vector and insert, are digested with NdeI and EcoRI restriction enzymes.
To create full-length DHX8 (fl-DHX8-His6) and DHX8Δ547-His6, PCR primers are designed, so that a His6-tag is fused to the C-terminus of DHX8. The resultant PCR amplicons encoding fl-DHX8 (M1 to R1220) or DHX8Δ547 (A548 to R1220) are inserted into the "pFBDM" vector[20] downstream of the polyhedrin promoter. Both, vector and insert, are digested with BamHI and NotI restriction enzymes.[9]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000067596 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000034931 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "DHX8 Gene - GeneCards | DHX8 Protein | DHX8 Antibody". www.genecards.org. Retrieved 2019-10-20.
- ↑ "DHX8 - ATP-dependent RNA helicase DHX8 - Homo sapiens (Human) - DHX8 gene & protein". UniProt. Retrieved 2019-10-20.
- ↑ English MA, Lei L, Blake T, Wincovitch SM, Sood R, Azuma M, et al. (May 2012). "Incomplete splicing, cell division defects, and hematopoietic blockage in dhx8 mutant zebrafish". Developmental Dynamics. 241 (5): 879–89. doi:10.1002/dvdy.23774. PMC 3328592. PMID 22411201.
- ↑ Dziuba N, Ferguson MR, O'Brien WA, Sanchez A, Prussia AJ, McDonald NJ, et al. (October 2012). "Identification of cellular proteins required for replication of human immunodeficiency virus type 1". AIDS Research and Human Retroviruses. 28 (10): 1329–39. doi:10.1089/aid.2011.0358. PMC 3448097. PMID 22404213.
- 1 2 3 4 5 6 PDB: 6HYU; Felisberto-Rodrigues C, Thomas JC, McAndrew C, Le Bihan YV, Burke R, Workman P, van Montfort RL (September 2019). "Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release". The Biochemical Journal. 476 (18): 2521–2543. doi:10.1042/BCJ20190383. PMID 31409651.
- ↑ Zhang X, Zhan X, Yan C, Zhang W, Liu D, Lei J, Shi Y (April 2019). "Structures of the human spliceosomes before and after release of the ligated exon". Cell Research. 29 (4): 274–285. doi:10.1038/s41422-019-0143-x. PMC 6461851. PMID 30728453.
- ↑ "Protein: DHX8_HUMAN (Q14562)". InterPro.
- ↑ "mRNA splicing, via spliceosome". Gene Ontology and GO Annotations. European Bioinformatics Institute (EMBL-EBI). Archived from the original on 2019-07-21.
- ↑ "Spliceosomal Complex Disassembly". Gene Ontology and GO Annotations. European Bioinformatics Institute (EMBL-EBI). Archived from the original on 2019-10-22.
- ↑ "ATP binding". Gene Ontology and GO Annotations. European Bioinformatics Institute (EMBL-EBI). Archived from the original on 2017-09-27.
- ↑ "RNA helicase activity". Gene Ontology and GO Annotations. European Bioinformatics Institute (EMBL-EBI). Archived from the original on 2019-08-14.
- ↑ "UniProtKB - Q14562 (DHX8_HUMAN)". UniProt.
- 1 2 "Tissue expression of DHX8 - Summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2019-10-25.
- ↑ Maity R, Pauty J, Krietsch J, Buisson R, Genois MM, Masson JY (October 2013). "GST-His purification: a two-step affinity purification protocol yielding full-length purified proteins". Journal of Visualized Experiments (80): e50320. doi:10.3791/50320. PMC 3964817. PMID 24193370.
- ↑ "Bac-to-Bac Vector Kit - Thermo Fisher Scientific". www.thermofisher.com. Retrieved 2019-10-24.
- ↑ "Addgene: pFBDM". Addgene. Retrieved 2019-10-24.