A DNA construct is an artificially-designed segment of DNA borne on a vector that can be used to incorporate genetic material into a target tissue or cell.[1] A DNA construct contains a DNA insert, called a transgene, delivered via a transformation vector which allows the insert sequence to be replicated and/or expressed in the target cell. This gene can be cloned from a naturally occurring gene,[2] or synthetically constructed.[3] The vector can be delivered using physical, chemical or viral methods.[4] Typically, the vectors used in DNA constructs contain an origin of replication, a multiple cloning site, and a selectable marker.[2] Certain vectors can carry additional regulatory elements based on the expression system involved.[5]
DNA constructs can be as small as a few thousand base pairs (kbp) of DNA carrying a single gene, using vectors such as plasmids or bacteriophages, or as large as hundreds of kbp for large-scale genomic studies using an artificial chromosome.[2] A DNA construct may express wildtype protein, prevent the expression of certain genes by expressing competitors or inhibitors, or express mutant proteins, such as deletion mutations or missense mutations. DNA constructs are widely adapted in molecular biology research for techniques such as DNA sequencing, protein expression, and RNA studies.[5]
History
The first standardized vector, pBR220, was designed in 1977 by researchers in Herbert Boyer’s lab. The plasmid contains various restriction enzyme sites and a stable antibiotic-resistance gene free from transposon activities.[6]
In 1982, Jeffrey Vieira and Joachim Messing described the development of M13mp7-derived pUC vectors that consist of a multiple cloning site and allow for more efficient sequencing and cloning using a set of universal M13 primers. Three years later, the currently popular pUC19 plasmid was engineered by the same scientists.[7]
Construction
The gene on a DNA sequence of interest can either be cloned from an existing sequence or developed synthetically. To clone a naturally occurring sequence in an organism, the organism's DNA is first cut with restriction enzymes, which recognize DNA sequences and cut them, around the target gene. The gene can then be amplified using polymerase chain reaction (PCR). Typically, this process includes using short sequences known as primers to initially hybridize to the target sequence; in addition, point mutations can be introduced in the primer sequences and then copied in each cycle in order to modify the target sequence.[2]
It is also possible to synthesize a target DNA strand for a DNA construct. Short strands of DNA known as oligonucleotides can be developed using column-based synthesis, in which bases are added one at a time to a strand of DNA attached to a solid phase. Each base has a protecting group to prevent linkage that is not removed until the next base is ready to be added, ensuring that they are linked in the correct sequence. Oligonucleotides can also be synthesized on a microarray, which allows for tens of thousands of sequences to be synthesized at once, in order to reduce cost.[3] To synthesize a larger gene, oligonucleotides are developed with overlapping sequences on the ends and then joined together. The most common method is called polymerase cycling assembly (PCA): fragments hybridize at the overlapping regions and are extended, and larger fragments are created in each cycle.[2]
Once a sequence has been isolated, it must be inserted into a vector. The easiest way to do this is to cut the vector DNA using restriction enzymes; if the same enzymes were used to isolate the target sequence, then the same "overhang" sequences will be created on each end allowing for hybridization. Once the target gene has hybridized to the vector DNA, they can be joined using a DNA ligase.[2] An alternative strategy uses recombination between homologous sites on the target gene and the vector sequence, eliminating the need for restriction enzymes.[8]
Modes of delivery
There are three general categories of DNA construct delivery: physical, chemical, and viral.[4] Physical methods, which deliver the DNA by physically penetrating the cell, include microinjection, electroporation, and biolistics.[9] Chemical methods rely on chemical reactions to deliver the DNA and include transformation with cells made competent using calcium phosphate as well as delivery via lipid nanoparticles.[10][11] Viral methods use a variety of viral vectors to deliver the DNA, including adenovirus, lentivirus, and herpes simplex virus[12]
Vector structure
In addition to the target gene, there are three important elements in a vector: an origin of replication, a selectable marker, and a multiple cloning site. An origin of replication is a DNA sequence that starts the process of DNA replication, allowing the vector to clone itself. A multiple cloning site contains binding sites for several restriction enzymes, making it easier to insert different DNA sequences into the vector. A selectable marker confers some trait that can be easily selected for in a host cell, so that it can be determined whether transformation was successful. The most common selectable markers are genes for antibiotic resistance, so that host cells without the construct will die off when exposed to the antibody and only host cells with the construct will remain.[2]
Types of DNA constructs
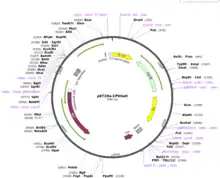
- Bacterial plasmids are circular sections of DNA that naturally replicate in bacteria.[2] Plasmids are capable of holding inserts up to approximately 20 kbp in length. These types of constructs typically contain a gene offering antibiotic-resistance, an origin of replication, regulatory elements such as Lac inhibitors, a polylinker, and a protein tag which facilitates protein purification.[14]
- Bacteriophage Vectors are viruses that can infect bacteria and replicate their own DNA.[2]
- Artificial chromosomes are commonly used in genome project studies due to their ability to hold inserts up to 350 kbp. These vectors are derived from the F plasmid, taking advantage of the high stability and conjugational ability introduced by the F factor.[15]
- Fosmids are a hybrid between bacterial F plasmids and λ phage cloning techniques. Inserts are pre-packaged into phage particles, then inserted into the host cell with the ability to hold ~45 kbp. They are typically used to generate a DNA library due to their increased stability.[16]
Applications
DNA constructs can be used to produce proteins, including both naturally occurring proteins and engineered mutant proteins. These proteins can be used to make therapeutic products, such as pharmaceuticals and antibodies. DNA constructs can also change the expression levels of other genes by expressing regulatory sequences such as promoters and inhibitors. Additionally, DNA constructs can be used for research such as creating genomic libraries, sequencing cloned DNA, and studying RNA and protein expression.[5]
See also
References
- ↑ Pinkert, Carl (2014). Transgenic animal technology: A laboratory handbook. Amsterdam: Elsevier. p. 692. ISBN 9780124095366.
- 1 2 3 4 5 6 7 8 9 Carter, Matt; Shieh, Jennifer C. (2010), "Molecular Cloning and Recombinant DNA Technology", Guide to Research Techniques in Neuroscience, Elsevier, pp. 207–227, doi:10.1016/b978-0-12-374849-2.00009-4, ISBN 978-0-12-374849-2, retrieved 2021-11-10
- 1 2 Hughes, Randall A.; Ellington, Andrew D. (January 2017). "Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology". Cold Spring Harbor Perspectives in Biology. 9 (1): a023812. doi:10.1101/cshperspect.a023812. ISSN 1943-0264. PMC 5204324. PMID 28049645.
- 1 2 Carter, Matt; Shieh, Jennifer C. (2010), "Gene Delivery Strategies", Guide to Research Techniques in Neuroscience, Elsevier, pp. 229–242, doi:10.1016/b978-0-12-374849-2.00010-0, ISBN 978-0-12-374849-2, retrieved 2020-10-24
- 1 2 3 Glick, Bernard R.; Patten, Cheryl L. (2017). Molecular Biotechnology: Principles and Applications of Recombinant DNA. Washington DC: ASM Press.
- ↑ Bolivar, Francisco; Rodriguez, Raymond L.; Betlach, Mary C.; Boyer, Herbert W. (1977-11-01). "Construction and characterization of new cloning vehicles I. Ampicillin-resistant derivatives of the plasmid pMB9". Gene. 2 (2): 75–93. doi:10.1016/0378-1119(77)90074-9. ISSN 0378-1119. PMID 344136.
- ↑ Yanisch-Perron, Celeste; Vieira, Jeffrey; Messing, Joachim (1985-01-01). "Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors". Gene. 33 (1): 103–119. doi:10.1016/0378-1119(85)90120-9. ISSN 0378-1119. PMID 2985470.
- ↑ Copeland, Neal G.; Jenkins, Nancy A.; Court, Donald L. (October 2001). "Recombineering: a powerful new tool for mouse functional genomics". Nature Reviews Genetics. 2 (10): 769–779. doi:10.1038/35093556. ISSN 1471-0064. PMID 11584293. S2CID 10916548.
- ↑ Mehierhumbert, S; Guy, R (2005-04-05). "Physical methods for gene transfer: Improving the kinetics of gene delivery into cells". Advanced Drug Delivery Reviews. 57 (5): 733–753. doi:10.1016/j.addr.2004.12.007. PMID 15757758.
- ↑ Felgner, P. L.; Gadek, T. R.; Holm, M.; Roman, R.; Chan, H. W.; Wenz, M.; Northrop, J. P.; Ringold, G. M.; Danielsen, M. (1987-11-01). "Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure". Proceedings of the National Academy of Sciences. 84 (21): 7413–7417. Bibcode:1987PNAS...84.7413F. doi:10.1073/pnas.84.21.7413. ISSN 0027-8424. PMC 299306. PMID 2823261.
- ↑ Kingston, Robert E.; Chen, Claudia A.; Rose, John K. (2003). "Calcium Phosphate Transfection". Current Protocols in Molecular Biology. 63 (1): 9.1.1–9.1.11. doi:10.1002/0471142727.mb0901s63. ISSN 1934-3647. PMID 18265332. S2CID 46188175.
- ↑ Robbins, Paul D.; Ghivizzani, Steven C. (1998). "Viral Vectors for Gene Therapy". Pharmacology & Therapeutics. 80 (1): 35–47. doi:10.1016/S0163-7258(98)00020-5. PMID 9804053.
- ↑ Shen, Aimee; Lupardus, Patrick J.; Morell, Montse; Ponder, Elizabeth L.; Sadaghiani, A. Masoud; Garcia, K. Christopher; Bogyo, Matthew (2009-12-02). Xu, Wenqing (ed.). "Simplified, Enhanced Protein Purification Using an Inducible, Autoprocessing Enzyme Tag". PLOS ONE. 4 (12): e8119. Bibcode:2009PLoSO...4.8119S. doi:10.1371/journal.pone.0008119. ISSN 1932-6203. PMC 2780291. PMID 19956581.
- ↑ Griffiths, Anthony J.F. (2015). Introduction To Genetic Analysis. New York: W.H. Freeman & Company. ISBN 978-1464188046.
- ↑ Godiska, R.; Wu, C. -C.; Mead, D. A. (2013-01-01), "Genomic Libraries", in Maloy, Stanley; Hughes, Kelly (eds.), Brenner's Encyclopedia of Genetics (Second Edition), San Diego: Academic Press, pp. 306–309, doi:10.1016/b978-0-12-374984-0.00641-0, ISBN 978-0-08-096156-9, retrieved 2020-11-06
- ↑ Hu, Bo; Khara, Pratick; Christie, Peter J. (2019-07-09). "Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli". Proceedings of the National Academy of Sciences. 116 (28): 14222–14227. Bibcode:2019PNAS..11614222H. doi:10.1073/pnas.1904428116. ISSN 0027-8424. PMC 6628675. PMID 31239340.