Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor, which can be converted to the kinds of tetrahydrofolate cofactors used in 1-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the DHFR gene.[5][6] It is found in the q14.1 region of chromosome 5.[7]
There are two structural classes of DHFR, evolutionarily unrelated to each other. The former is usually just called DHFR and is found in bacterial chromosomes and animals. In bacteria, however, antibiotic pressure has caused this class to evolve different patterns of binding diaminoheterocyclic molecules, leading to many "types" named under this class, while mammalian ones remain highly similar.[8] The latter (type II), represented by the plastid-encoded R67, is a tiny enzyme that works by forming a homotetramer.[9]
Function
| Dihydrofolate reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 1.5.1.3 | ||||||||
| CAS no. | 9002-03-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Dihydrofolate reductase converts dihydrofolate into tetrahydrofolate, a proton shuttle required for the de novo synthesis of purines, thymidylic acid, and certain amino acids. While the functional dihydrofolate reductase gene has been mapped to chromosome 5, multiple intronless processed pseudogenes or dihydrofolate reductase-like genes have been identified on separate chromosomes.[10]
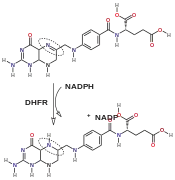 Reaction catalyzed by DHFR.
Reaction catalyzed by DHFR.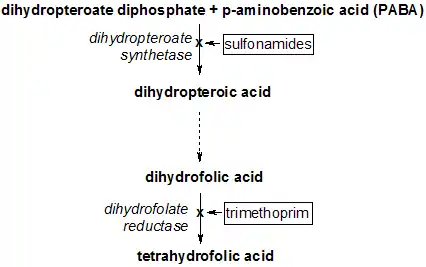 Tetrahydrofolate synthesis pathway.
Tetrahydrofolate synthesis pathway.
Found in all organisms, DHFR has a critical role in regulating the amount of tetrahydrofolate in the cell. Tetrahydrofolate and its derivatives are essential for purine and thymidylate synthesis, which are important for cell proliferation and cell growth.[11] DHFR plays a central role in the synthesis of nucleic acid precursors, and it has been shown that mutant cells that completely lack DHFR require glycine, a purine, and thymidine to grow.[12] DHFR has also been demonstrated as an enzyme involved in the salvage of tetrahydrobiopterin from dihydrobiopterin[13]
Structure
| Dihydrofolate reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of chicken liver dihydrofolate reductase. PDB entry 8dfr | |||||||||
| Identifiers | |||||||||
| Symbol | DHFR_1 | ||||||||
| Pfam | PF00186 | ||||||||
| Pfam clan | CL0387 | ||||||||
| InterPro | IPR001796 | ||||||||
| PROSITE | PDOC00072 | ||||||||
| SCOP2 | 1dhi / SCOPe / SUPFAM | ||||||||
| |||||||||
A central eight-stranded beta-pleated sheet makes up the main feature of the polypeptide backbone folding of DHFR.[14] Seven of these strands are parallel and the eighth runs antiparallel. Four alpha helices connect successive beta strands.[15] Residues 9 – 24 are termed "Met20" or "loop 1" and, along with other loops, are part of the major subdomain that surround the active site.[16] The active site is situated in the N-terminal half of the sequence, which includes a conserved Pro-Trp dipeptide; the tryptophan has been shown to be involved in the binding of substrate by the enzyme.[15]
Mechanism
General mechanism

DHFR catalyzes the transfer of a hydride from NADPH to dihydrofolate with an accompanying protonation to produce tetrahydrofolate.[11] In the end, dihydrofolate is reduced to tetrahydrofolate and NADPH is oxidized to NADP+. The high flexibility of Met20 and other loops near the active site play a role in promoting the release of the product, tetrahydrofolate. In particular the Met20 loop helps stabilize the nicotinamide ring of the NADPH to promote the transfer of the hydride from NADPH to dihydrofolate.[16]
The mechanism of this enzyme is stepwise and steady-state random. Specifically, the catalytic reaction begins with the NADPH and the substrate attaching to the binding site of the enzyme, followed by the protonation and the hydride transfer from the cofactor NADPH to the substrate. However, two latter steps do not take place simultaneously in a same transition state.[17][18] In a study using computational and experimental approaches, Liu et al conclude that the protonation step precedes the hydride transfer.[19]
.png.webp)
DHFR's enzymatic mechanism is shown to be pH dependent, particularly the hydride transfer step, since pH changes are shown to have remarkable influence on the electrostatics of the active site and the ionization state of its residues.[19] The acidity of the targeted nitrogen on the substrate is important in the binding of the substrate to the enzyme's binding site which is proved to be hydrophobic even though it has direct contact to water.[17][20] Asp27 is the only charged hydrophilic residue in the binding site, and neutralization of the charge on Asp27 may alter the pKa of the enzyme. Asp27 plays a critical role in the catalytic mechanism by helping with protonation of the substrate and restraining the substrate in the conformation favorable for the hydride transfer.[21][17][20] The protonation step is shown to be associated with enol tautomerization even though this conversion is not considered favorable for the proton donation.[18] A water molecule is proved to be involved in the protonation step.[22][23][24] Entry of the water molecule to the active site of the enzyme is facilitated by the Met20 loop.[25]
Conformational changes of DHFR

The catalytic cycle of the reaction catalyzed by DHFR incorporates five important intermediate: holoenzyme (E:NADPH), Michaelis complex (E:NADPH:DHF), ternary product complex (E:NADP+:THF), tetrahydrofolate binary complex (E:THF), and THF‚NADPH complex (E:NADPH:THF). The product (THF) dissociation step from E:NADPH:THF to E:NADPH is the rate determining step during steady-state turnover.[21]
Conformational changes are critical in DHFR's catalytic mechanism.[26] The Met20 loop of DHFR is able to open, close or occlude the active site.[23][17] Correspondingly, three different conformations classified as the opened, closed and occluded states are assigned to Met20. In addition, an extra distorted conformation of Met20 was defined due to its indistinct characterization results.[23] The Met20 loop is observed in its occluded conformation in the three product ligating intermediates, where the nicotinamide ring is occluded from the active site. This conformational feature accounts for the fact that the substitution of NADP+ by NADPH is prior to product dissociation. Thus, the next round of reaction can occur upon the binding of substrate.[21]
R67 DHFR
| R67 dihydrofolate reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 R67 in complex with DHF and NADP+, monomer. PDB entry 2rk1. | |||||||||
| Identifiers | |||||||||
| Symbol | DHFR_2 | ||||||||
| Pfam | PF06442 | ||||||||
| InterPro | IPR009159 | ||||||||
| SCOP2 | 1vif / SCOPe / SUPFAM | ||||||||
| |||||||||
Due to its unique structure and catalytic features, R67 DHFR is widely studied. R67 DHFR is a type II R-plasmid-encoded DHFR without geneticay or structural relation to the E. coli chromosomal DHFR. It is a homotetramer that possesses the 222 symmetry with a single active site pore that is exposed to solvent.[27] This symmetry of active site results in the different binding mode of the enzyme: It can bind with two dihydrofolate (DHF) molecules with positive cooperativity or two NADPH molecules with negative cooperativity, or one substrate plus one, but only the latter one has the catalytical activity.[28] Compare with E. coli chromosomal DHFR, it has higher Km in binding dihydrofolate (DHF) and NADPH. The much lower catalytical kinetics show that hydride transfer is the rate determine step rather than product (THF) release.[29]
In the R67 DHFR structure, the homotetramer forms an active site pore. In the catalytical process, DHF and NADPH enters into the pore from opposite position. The π-π stacking interaction between NADPH's nicotinamide ring and DHF's pteridine ring tightly connect two reactants in the active site. However, the flexibility of p-aminobenzoylglutamate tail of DHF was observed upon binding which can promote the formation of the transition state.[30]
 Reaction Kinetics comparison between E. coli DHFR (EcDHFR) and R67 DHFR
Reaction Kinetics comparison between E. coli DHFR (EcDHFR) and R67 DHFR Structure difference of substrate binding in EcDHFR and R67 DHFR
Structure difference of substrate binding in EcDHFR and R67 DHFR
Clinical significance
DHFR mutations cause dihydrofolate reductase deficiency, a rare autosomal recessive inborn error of folate metabolism that results in megaloblastic anemia, pancytopenia and severe cerebral folate deficiency. These issues can be overcome by supplementation with a reduced form of folate, usually folinic acid.[10][31][32]
Therapeutic applications
DHFR is an attractive pharmaceutical target for inhibition due to its pivotal role in DNA precursor (thymine) synthesis. Trimethoprim, an antibiotic, inhibits bacterial DHFR while methotrexate, a chemotherapy agent, inhibits mammalian DHFR. However, resistance has developed against some drugs, as a result of mutational changes in DHFR itself.[33]
Cancer
DHFR is responsible for the levels of tetrahydrofolate in a cell, and the inhibition of DHFR can limit the growth and proliferation of cells that are characteristic of cancer and bacterial infections. Methotrexate, a competitive inhibitor of DHFR, is one such anticancer drug that inhibits DHFR.[34]
Folate is necessary for growth,[35] and the pathway of the metabolism of folate is a target in developing treatments for cancer. DHFR is one such target. A regimen of fluorouracil, doxorubicin, and methotrexate was shown to prolong survival in patients with advanced gastric cancer.[36] Further studies into inhibitors of DHFR can lead to more ways to treat cancer.
Infection
Bacteria also need DHFR to grow and multiply and hence inhibitors selective for bacterial DHFR have found application as antibacterial agents.[37] Trimethoprim has shown to have activity against a variety of Gram-positive bacterial pathogens.[37] However, resistance to trimethoprim and other drugs aimed at DHFR can arise due to a variety of mechanisms, limiting the success of their therapeutical uses.[38][39][40] Resistance can arise from DHFR gene amplification, mutations in DHFR,[41][42] decrease in the uptake of the drugs, among others. Regardless, trimethoprim and sulfamethoxazole in combination has been used as an antibacterial agent for decades.[37]
Pyrimethamine is a widely used antiprotozoal agent.[43]
Other classes of compounds that target DHFR in general, and bacterial DHFRs in particular, belong to the classes such as diaminopteridines, diaminotriazines, diaminopyrroloquinazolines, stilbenes, chalcones, deoxybenzoins, diaminoquinazolines, diaminopyrroloquinazolines, to name but a few.
Potential anthrax treatment

Dihydrofolate reductase from Bacillus anthracis (BaDHFR) is a validated drug target in the treatment of the infectious disease, anthrax. BaDHFR is less sensitive to trimethoprim analogs than is dihydrofolate reductase from other species such as Escherichia coli, Staphylococcus aureus, and Streptococcus pneumoniae. A structural alignment of dihydrofolate reductase from all four species shows that only BaDHFR has the combination phenylalanine and tyrosine in positions 96 and 102, respectively.
BaDHFR's resistance to trimethoprim analogs is due to these two residues (F96 and Y102), which also confer improved kinetics and catalytic efficiency.[44] Current research uses active site mutants in BaDHFR to guide lead optimization for new antifolate inhibitors.[44]
As a research tool
DHFR has been used as a tool to detect protein–protein interactions in a protein-fragment complementation assay (PCA), using a split-protein approach.[45]
DHFR-lacking CHO cells are the most commonly used cell line for the production of recombinant proteins. These cells are transfected with a plasmid carrying the dhfr gene and the gene for the recombinant protein in a single expression system, and then subjected to selective conditions in thymidine-lacking medium. Only the cells with the exogenous DHFR gene along with the gene of interest survive. Supplementation of this medium with methotrexate, a competitive inhibitor of DHFR, can further select for those cells expressing the highest levels of DHFR, and thus, select for the top recombinant protein producers.[46]
Interactions
Dihydrofolate reductase has been shown to interact with GroEL[47] and Mdm2.[48]
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ↑ The interactive pathway map can be edited at WikiPathways: "FluoropyrimidineActivity_WP1601".
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000228716 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000021707 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ Chen MJ, Shimada T, Moulton AD, Harrison M, Nienhuis AW (December 1982). "Intronless human dihydrofolate reductase genes are derived from processed RNA molecules". Proceedings of the National Academy of Sciences of the United States of America. 79 (23): 7435–9. Bibcode:1982PNAS...79.7435C. doi:10.1073/pnas.79.23.7435. PMC 347354. PMID 6961421.
- ↑ Chen MJ, Shimada T, Moulton AD, Cline A, Humphries RK, Maizel J, Nienhuis AW (March 1984). "The functional human dihydrofolate reductase gene". The Journal of Biological Chemistry. 259 (6): 3933–43. doi:10.1016/S0021-9258(17)43186-3. PMID 6323448.
- ↑ "DHFR dihydrofolate reductase [Homo sapiens (human)]". Gene - NCBI. Retrieved 21 February 2023.
- ↑ Smith SL, Patrick P, Stone D, Phillips AW, Burchall JJ (November 1979). "Porcine liver dihydrofolate reductase. Purification, properties, and amino acid sequence". The Journal of Biological Chemistry. 254 (22): 11475–84. doi:10.1016/S0021-9258(19)86510-9. PMID 500653.
- ↑ Krahn JM, Jackson MR, DeRose EF, Howell EE, London RE (25 December 2007). "Crystal structure of a type II dihydrofolate reductase catalytic ternary complex". Biochemistry. 46 (51): 14878–88. doi:10.1021/bi701532r. PMC 3743094. PMID 18052202.
- 1 2 "Entrez Gene: DHFR dihydrofolate reductase".
- 1 2 Schnell JR, Dyson HJ, Wright PE (2004). "Structure, dynamics, and catalytic function of dihydrofolate reductase". Annual Review of Biophysics and Biomolecular Structure. 33 (1): 119–40. doi:10.1146/annurev.biophys.33.110502.133613. PMID 15139807. S2CID 28611812.
- ↑ Urlaub G, Chasin LA (July 1980). "Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity". Proceedings of the National Academy of Sciences of the United States of America. 77 (7): 4216–20. Bibcode:1980PNAS...77.4216U. doi:10.1073/pnas.77.7.4216. PMC 349802. PMID 6933469.
- ↑ Crabtree MJ, Tatham AL, Hale AB, Alp NJ, Channon KM (October 2009). "Critical role for tetrahydrobiopterin recycling by dihydrofolate reductase in regulation of endothelial nitric-oxide synthase coupling: relative importance of the de novo biopterin synthesis versus salvage pathways". The Journal of Biological Chemistry. 284 (41): 28128–36. doi:10.1074/jbc.M109.041483. PMC 2788863. PMID 19666465.
- ↑ Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K (July 1977). "Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate". Science. 197 (4302): 452–5. Bibcode:1977Sci...197..452M. doi:10.1126/science.17920. PMID 17920.
- 1 2 Filman DJ, Bolin JT, Matthews DA, Kraut J (November 1982). "Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 Å resolution. II. Environment of bound NADPH and implications for catalysis". The Journal of Biological Chemistry. 257 (22): 13663–72. doi:10.1016/S0021-9258(18)33498-7. PMID 6815179.
- 1 2 Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE (August 2001). "Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism". Biochemistry. 40 (33): 9846–59. doi:10.1021/bi010621k. PMID 11502178.
- 1 2 3 4 Rod TH, Brooks CL (July 2003). "How dihydrofolate reductase facilitates protonation of dihydrofolate". Journal of the American Chemical Society. 125 (29): 8718–9. doi:10.1021/ja035272r. PMID 12862454.
- 1 2 Wan Q, Bennett BC, Wilson MA, Kovalevsky A, Langan P, Howell EE, Dealwis C (December 2014). "Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography". Proceedings of the National Academy of Sciences of the United States of America. 111 (51): 18225–30. Bibcode:2014PNAS..11118225W. doi:10.1073/pnas.1415856111. PMC 4280638. PMID 25453083.
- 1 2 Liu CT, Francis K, Layfield JP, Huang X, Hammes-Schiffer S, Kohen A, Benkovic SJ (December 2014). "Escherichia coli dihydrofolate reductase catalyzed proton and hydride transfers: temporal order and the roles of Asp27 and Tyr100". Proceedings of the National Academy of Sciences of the United States of America. 111 (51): 18231–6. Bibcode:2014PNAS..11118231L. doi:10.1073/pnas.1415940111. PMC 4280594. PMID 25453098.
- 1 2 Czekster CM, Vandemeulebroucke A, Blanchard JS (January 2011). "Kinetic and chemical mechanism of the dihydrofolate reductase from Mycobacterium tuberculosis". Biochemistry. 50 (3): 367–75. doi:10.1021/bi1016843. PMC 3074011. PMID 21138249.
- 1 2 3 Fierke CA, Johnson KA, Benkovic SJ (June 1987). "Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli". Biochemistry. 26 (13): 4085–92. doi:10.1021/bi00387a052. PMID 3307916.
- ↑ Reyes VM, Sawaya MR, Brown KA, Kraut J (February 1995). "Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications". Biochemistry. 34 (8): 2710–23. doi:10.1021/bi00008a039. PMID 7873554.
- 1 2 3 Sawaya MR, Kraut J (January 1997). "Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence". Biochemistry. 36 (3): 586–603. doi:10.1021/bi962337c. PMID 9012674.
- ↑ Chen YQ, Kraut J, Blakley RL, Callender R (June 1994). "Determination by Raman spectroscopy of the pKa of N5 of dihydrofolate bound to dihydrofolate reductase: mechanistic implications". Biochemistry. 33 (23): 7021–6. doi:10.1021/bi00189a001. PMID 8003467.
- ↑ Shrimpton P, Allemann RK (June 2002). "Role of water in the catalytic cycle of E. coli dihydrofolate reductase". Protein Science. 11 (6): 1442–51. doi:10.1110/ps.5060102. PMC 2373639. PMID 12021443.
- ↑ Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG (December 2005). "Conformation coupled enzyme catalysis: single-molecule and transient kinetics investigation of dihydrofolate reductase". Biochemistry. 44 (51): 16835–43. doi:10.1021/bi051378i. PMID 16363797.
- ↑ Narayana N, Matthews DA, Howell EE, Nguyen-huu X (November 1995). "A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site". Nature Structural Biology. 2 (11): 1018–25. doi:10.1038/nsb1195-1018. PMID 7583655. S2CID 11914241.
- ↑ Bradrick TD, Beechem JM, Howell EE (September 1996). "Unusual binding stoichiometries and cooperativity are observed during binary and ternary complex formation in the single active pore of R67 dihydrofolate reductase, a D2 symmetric protein". Biochemistry. 35 (35): 11414–24. doi:10.1021/bi960205d. PMID 8784197.
- ↑ Park H, Zhuang P, Nichols R, Howell EE (January 1997). "Mechanistic studies of R67 dihydrofolate reductase. Effects of pH and an H62C mutation". The Journal of Biological Chemistry. 272 (4): 2252–8. doi:10.1074/jbc.272.4.2252. PMID 8999931.
- ↑ Kamath G, Howell EE, Agarwal PK (October 2010). "The tail wagging the dog: insights into catalysis in R67 dihydrofolate reductase". Biochemistry. 49 (42): 9078–88. doi:10.1021/bi1007222. PMID 20795731.
- ↑ Banka S, Blom HJ, Walter J, Aziz M, Urquhart J, Clouthier CM, et al. (February 2011). "Identification and characterization of an inborn error of metabolism caused by dihydrofolate reductase deficiency". American Journal of Human Genetics. 88 (2): 216–25. doi:10.1016/j.ajhg.2011.01.004. PMC 3035707. PMID 21310276.
- ↑ Nyhan WL, Hoffmann GF, Barshop BA (30 December 2011). Atlas of Inherited Metabolic Diseases 3E. CRC Press. pp. 141–. ISBN 978-1-4441-4948-7.
{{cite book}}: CS1 maint: overridden setting (link) - ↑ Cowman AF, Lew AM (November 1989). "Antifolate drug selection results in duplication and rearrangement of chromosome 7 in Plasmodium chabaudi". Molecular and Cellular Biology. 9 (11): 5182–8. doi:10.1128/mcb.9.11.5182. PMC 363670. PMID 2601715.
- ↑ Li R, Sirawaraporn R, Chitnumsub P, Sirawaraporn W, Wooden J, Athappilly F, Turley S, Hol WG (January 2000). "Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs". Journal of Molecular Biology. 295 (2): 307–23. doi:10.1006/jmbi.1999.3328. PMID 10623528. S2CID 24527344.
- ↑ Bailey SW, Ayling JE (September 2009). "The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake". Proceedings of the National Academy of Sciences of the United States of America. 106 (36): 15424–9. doi:10.1073/pnas.0902072106. PMC 2730961. PMID 19706381.
- ↑ Murad AM, Santiago FF, Petroianu A, Rocha PR, Rodrigues MA, Rausch M (July 1993). "Modified therapy with 5-fluorouracil, doxorubicin, and methotrexate in advanced gastric cancer". Cancer. 72 (1): 37–41. doi:10.1002/1097-0142(19930701)72:1<37::AID-CNCR2820720109>3.0.CO;2-P. PMID 8508427.
- 1 2 3 Hawser S, Lociuro S, Islam K (March 2006). "Dihydrofolate reductase inhibitors as antibacterial agents". Biochemical Pharmacology. 71 (7): 941–8. doi:10.1016/j.bcp.2005.10.052. PMID 16359642.
- ↑ Narayana N, Matthews DA, Howell EE, Nguyen-huu X (November 1995). "A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site". Nature Structural Biology. 2 (11): 1018–25. doi:10.1038/nsb1195-1018. PMID 7583655. S2CID 11914241.
- ↑ Huennekens FM (June 1996). "In search of dihydrofolate reductase". Protein Science. 5 (6): 1201–8. doi:10.1002/pro.5560050626. PMC 2143423. PMID 8762155.
- ↑ Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR (July 2002). "Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1587 (2–3): 164–73. doi:10.1016/S0925-4439(02)00079-0. PMID 12084458.
- ↑ Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R (December 2011). "Evolutionary paths to antibiotic resistance under dynamically sustained drug selection". Nature Genetics. 44 (1): 101–5. doi:10.1038/ng.1034. PMC 3534735. PMID 22179135.
- ↑ Rodrigues JV, Bershtein S, Li A, Lozovsky ER, Hartl DL, Shakhnovich EI (March 2016). "Biophysical principles predict fitness landscapes of drug resistance". Proceedings of the National Academy of Sciences of the United States of America. 113 (11): E1470-8. Bibcode:2016PNAS..113E1470R. doi:10.1073/pnas.1601441113. PMC 4801265. PMID 26929328.
- ↑ Benkovic SJ, Fierke CA, Naylor AM (March 1988). "Insights into enzyme function from studies on mutants of dihydrofolate reductase". Science. 239 (4844): 1105–10. Bibcode:1988Sci...239.1105B. doi:10.1126/science.3125607. PMID 3125607.
- 1 2 Beierlein JM, Karri NG, Anderson AC (October 2010). "Targeted mutations of Bacillus anthracis dihydrofolate reductase condense complex structure−activity relationships". Journal of Medicinal Chemistry. 53 (20): 7327–36. doi:10.1021/jm100727t. PMC 3618964. PMID 20882962.
- ↑ Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW (June 2008). "An in vivo map of the yeast protein interactome" (PDF). Science. 320 (5882): 1465–70. Bibcode:2008Sci...320.1465T. doi:10.1126/science.1153878. PMID 18467557. S2CID 1732896.
- ↑ Ng SK (2012). Generation of High-Expressing Cells by Methotrexate Amplification of Destabilized Dihydrofolate Reductase Selection Marker. Methods in Molecular Biology. Vol. 801. pp. 161–172. doi:10.1007/978-1-61779-352-3_11. ISBN 978-1-61779-351-6. PMID 21987253.
{{cite book}}:|journal=ignored (help) - ↑ Mayhew M, da Silva AC, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU (February 1996). "Protein folding in the central cavity of the GroEL-GroES chaperonin complex". Nature. 379 (6564): 420–6. Bibcode:1996Natur.379..420M. doi:10.1038/379420a0. PMID 8559246. S2CID 4310511.
- ↑ Maguire M, Nield PC, Devling T, Jenkins RE, Park BK, Polański R, Vlatković N, Boyd MT (May 2008). "MDM2 regulates dihydrofolate reductase activity through monoubiquitination". Cancer Research. 68 (9): 3232–42. doi:10.1158/0008-5472.CAN-07-5271. PMC 3536468. PMID 18451149.
Further reading
- Joska TM, Anderson AC (October 2006). "Structure-activity relationships of Bacillus cereus and Bacillus anthracis dihydrofolate reductase: toward the identification of new potent drug leads". Antimicrobial Agents and Chemotherapy. 50 (10): 3435–43. doi:10.1128/AAC.00386-06. PMC 1610094. PMID 17005826.
- Chan DC, Fu H, Forsch RA, Queener SF, Rosowsky A (June 2005). "Design, synthesis, and antifolate activity of new analogues of piritrexim and other diaminopyrimidine dihydrofolate reductase inhibitors with omega-carboxyalkoxy or omega-carboxy-1-alkynyl substitution in the side chain". Journal of Medicinal Chemistry. 48 (13): 4420–31. doi:10.1021/jm0581718. PMID 15974594.
- Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR (July 2002). "Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1587 (2–3): 164–73. doi:10.1016/S0925-4439(02)00079-0. PMID 12084458.
- Stockman BJ, Nirmala NR, Wagner G, Delcamp TJ, DeYarman MT, Freisheim JH (January 1992). "Sequence-specific 1H and 15N resonance assignments for human dihydrofolate reductase in solution". Biochemistry. 31 (1): 218–29. doi:10.1021/bi00116a031. PMID 1731871.
- Beltzer JP, Spiess M (December 1991). "In vitro binding of the asialoglycoprotein receptor to the beta adaptin of plasma membrane coated vesicles". The EMBO Journal. 10 (12): 3735–42. doi:10.1002/j.1460-2075.1991.tb04942.x. PMC 453108. PMID 1935897.
- Davies JF, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J (October 1990). "Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate". Biochemistry. 29 (40): 9467–79. doi:10.1021/bi00492a021. PMID 2248959.
- Will CL, Dolnick BJ (December 1989). "5-Fluorouracil inhibits dihydrofolate reductase precursor mRNA processing and/or nuclear mRNA stability in methotrexate-resistant KB cells". The Journal of Biological Chemistry. 264 (35): 21413–21. doi:10.1016/S0021-9258(19)30096-1. PMID 2592384.
- Masters JN, Attardi G (March 1985). "Discrete human dihydrofolate reductase gene transcripts present in polysomal RNA map with their 5' ends several hundred nucleotides upstream of the main mRNA start site". Molecular and Cellular Biology. 5 (3): 493–500. doi:10.1128/mcb.5.3.493. PMC 366741. PMID 2859520.
- Miszta H, Dabrowski Z, Lanotte M (November 1988). "In vitro patterns of enzymic tetrahydrofolate dehydrogenase (EC 1.5.1.3) expression in bone marrow stromal cells". Leukemia. 2 (11): 754–9. PMID 3185016.
- Oefner C, D'Arcy A, Winkler FK (June 1988). "Crystal structure of human dihydrofolate reductase complexed with folate". European Journal of Biochemistry. 174 (2): 377–85. doi:10.1111/j.1432-1033.1988.tb14108.x. PMID 3383852.
- Yang JK, Masters JN, Attardi G (June 1984). "Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5' non-coding sequence and strong intron size divergence from homologous mammalian genes". Journal of Molecular Biology. 176 (2): 169–87. doi:10.1016/0022-2836(84)90419-4. PMID 6235374.
- Masters JN, Yang JK, Cellini A, Attardi G (June 1983). "A human dihydrofolate reductase pseudogene and its relationship to the multiple forms of specific messenger RNA". Journal of Molecular Biology. 167 (1): 23–36. doi:10.1016/S0022-2836(83)80032-1. PMID 6306253.
- Chen MJ, Shimada T, Moulton AD, Cline A, Humphries RK, Maizel J, Nienhuis AW (March 1984). "The functional human dihydrofolate reductase gene". The Journal of Biological Chemistry. 259 (6): 3933–43. doi:10.1016/S0021-9258(17)43186-3. PMID 6323448.
- Funanage VL, Myoda TT, Moses PA, Cowell HR (October 1984). "Assignment of the human dihydrofolate reductase gene to the q11----q22 region of chromosome 5". Molecular and Cellular Biology. 4 (10): 2010–6. doi:10.1128/mcb.4.10.2010. PMC 369017. PMID 6504041.
- Masters JN, Attardi G (1983). "The nucleotide sequence of the cDNA coding for the human dihydrofolic acid reductase". Gene. 21 (1–2): 59–63. doi:10.1016/0378-1119(83)90147-6. PMID 6687716.
- Morandi C, Masters JN, Mottes M, Attardi G (April 1982). "Multiple forms of human dihydrofolate reductase messenger RNA. Cloning and expression in Escherichia coli of their DNA coding sequence". Journal of Molecular Biology. 156 (3): 583–607. doi:10.1016/0022-2836(82)90268-6. PMID 6750132.
- Bonifaci N, Sitia R, Rubartelli A (September 1995). "Nuclear translocation of an exogenous fusion protein containing HIV Tat requires unfolding". AIDS. 9 (9): 995–1000. doi:10.1097/00002030-199509000-00003. PMID 8527095. S2CID 8417864.
- Mayhew M, da Silva AC, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU (February 1996). "Protein folding in the central cavity of the GroEL-GroES chaperonin complex". Nature. 379 (6564): 420–6. Bibcode:1996Natur.379..420M. doi:10.1038/379420a0. PMID 8559246. S2CID 4310511.
- Gross M, Robinson CV, Mayhew M, Hartl FU, Radford SE (December 1996). "Significant hydrogen exchange protection in GroEL-bound DHFR is maintained during iterative rounds of substrate cycling". Protein Science. 5 (12): 2506–13. doi:10.1002/pro.5560051213. PMC 2143321. PMID 8976559.
- Schleiff E, Shore GC, Goping IS (March 1997). "Human mitochondrial import receptor, Tom20p. Use of glutathione to reveal specific interactions between Tom20-glutathione S-transferase and mitochondrial precursor proteins". FEBS Letters. 404 (2–3): 314–8. doi:10.1016/S0014-5793(97)00145-2. PMID 9119086. S2CID 29177508.
- Cody V, Galitsky N, Luft JR, Pangborn W, Rosowsky A, Blakley RL (November 1997). "Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523". Biochemistry. 36 (45): 13897–903. doi:10.1021/bi971711l. PMID 9374868.
- Vanguri VK, Wang S, Godyna S, Ranganathan S, Liau G (April 2000). "Thrombospondin-1 binds to polyhistidine with high affinity and specificity". The Biochemical Journal. 347 (Pt 2): 469–73. doi:10.1042/0264-6021:3470469. PMC 1220979. PMID 10749676.
External links
- 1988 Nobel lecture in Medicine
- Proteopedia: Dihydrofolate reductase
- Overview of all the structural information available in the PDB for UniProt: P00374 (Dihydrofolate reductase) at the PDBe-KB.

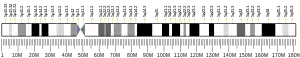








![1kms: HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9439), A LIPOPHILIC ANTIFOLATE](../I/PDB_1kms_EBI.jpg.webp)
![1kmv: HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9662), A LIPOPHILIC ANTIFOLATE](../I/PDB_1kmv_EBI.jpg.webp)
![1mvs: Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase](../I/PDB_1mvs_EBI.jpg.webp)
![1mvt: Analysis of Two Polymorphic Forms of a Pyrido[2,3-d]pyrimidine N9-C10 Reverse-Bridge Antifolate Binary Complex with Human Dihydrofolate Reductase](../I/PDB_1mvt_EBI.jpg.webp)