GCaMP is a genetically encoded calcium indicator (GECI) initially developed in 2001 by Junichi Nakai.[1] It is a synthetic fusion of green fluorescent protein (GFP), calmodulin (CaM), and M13, a peptide sequence from myosin light-chain kinase.[2] When bound to Ca2+, GCaMP fluoresces green with a peak excitation wavelength of 480 nm and a peak emission wavelength of 510 nm.[3] It is used in biological research to measure intracellular Ca2+ levels both in vitro and in vivo using virally transfected or transgenic cell and animal lines.[2][4] The genetic sequence encoding GCaMP can be inserted under the control of promoters exclusive to certain cell types, allowing for cell-type specific expression of GCaMP.[5] Since Ca2+ is a second messenger that contributes to many cellular mechanisms and signaling pathways, GCaMP allows researchers to quantify the activity of Ca2+-based mechanisms and study the role of Ca2+ ions in biological processes of interest.
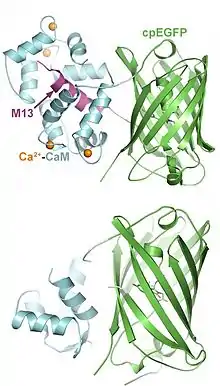
Structure
GCaMP consists of three key domains: an M13 domain at the N-terminus, a calmodulin (CaM) domain at the C-terminus, and a GFP domain in the center. The GFP domain is circularly permuted such that the native N- and C-termini are fused together by a six-amino-acid linking sequence, and the GFP sequence is split in the middle, creating new N- and C-termini that connect to the M13 and CaM domains.[6]
In the absence of Ca2+, the GFP chromophore is exposed to water and exists in a protonated state with minimal fluorescence intensity. Upon Ca2+ binding, the CaM domain undergoes a conformational change and tightly binds to the M13 domain alpha helix, preventing water molecules from accessing the chromophore. As a result, the chromophore rapidly deprotonates and converts into an anionic form that fluoresces brightly, similar to native GFP.[7]
History and development
In 2001, Nakai et al. reported the development of GCaMP1 as a Ca2+ probe with improved signal-to-noise ratio compared to previously developed fluorescent Ca2+ probes.[1] The first transgenic mouse expressing GCaMP1 was reported in 2004.[5] However, at 37 ˚C (physiological temperature in mammals), GCaMP1 did not fold stably or fluoresce, limiting its potential use as a calcium indicator in vivo.[1][8]
In 2006, Tallini et al. subsequently reported the improvement of GCaMP1 to GCaMP2, which exhibited brighter fluorescence than GCaMP1 and greater stability at mammalian body temperatures. Tallini et al. expressed GCaMP2 in cardiomyocytes in mouse embryos to perform the first in vivo GCaMP imaging of Ca2+ in mammals.[8]
Further modifications of GCaMP, including GCaMP3, GCaMP5, GCaMP6, and jGCaMP7, have been developed to progressively improve the signal, sensitivity, and dynamic range of Ca2+ detection,[2][9][10][11] with recent versions exhibiting fluorescence similar to native GFP.[11]
Variants in use
Both slow variants (GCaMP6s, jGCaMP7s) and fast variants (GCaMP6f, jGCaMP7f) are used in biological and neuroscience research. The slow variants are brighter and more sensitive to small changes in Ca2+ levels, such as single action potentials; on the other hand, the fast variants are less sensitive but respond more quickly, making them useful for tracking changes in Ca2+ levels over precise timescales.[12][13] GCaMP6 also has a medium variant, GCaMP6m, whose kinetics are intermediate between GCaMP6s and GCaMP6f.[12] Other variants of jGCaMP7 are also employed: jGCaMP7b exhibits bright baseline fluorescence and is used for imaging dendrites and axons, while jGCaMP7c exhibits greater contrast between maximal and baseline fluorescence and is advantageous for imaging large populations of neurons.[12]
In 2018, Yang et al. reported the development of GCaMP-X, generated by the addition of a calmodulin-binding motif. Since the GCaMP calmodulin domain, when unbound, disrupts L-type calcium channel gating, the added calmodulin-binding motif prevents GCaMP-X from interfering with calcium-dependent signaling mechanisms.[14]
In 2020, Zhang et al. reported the development of jGCaMP8, including sensitive, medium, and fast variants, which exhibit faster kinetics and greater sensitivity than the corresponding jGCaMP7 variants.[15]
Red fluorescent indicators have also been developed: jRCaMP1a and jRCaMP1b use a circular permutation of the red fluorescent protein mRuby instead of GFP, while jRGECO1a is based on the red fluorescent protein mApple.[12][16] Since the blue light used to excite GCaMP is scattered by tissue and the emitted green light is absorbed by blood, red fluorescent indicators provide more penetration and imaging depth in vivo than GCaMP. Use of red fluorescent indicators also avoids the photodamage caused by blue excitation light.[16] Moreover, red fluorescent indicators allow for concurrent use of optogenetics, which is difficult with GCaMP because the excitation wavelengths of GCaMP overlap with those of channelrhodopsin-2 (ChR2).[16][17] Simultaneous use of red and green GECIs can provide two-color visualization of different subcellular regions or cell populations.[16][17][18]
Applications in research
Neuronal activity
In neurons, action potentials induce neurotransmitter release at axon terminals by opening voltage-gated Ca2+ channels, allowing for Ca2+ influx. As a result, GCaMP is commonly used to measure increases in intracellular Ca2+ in neurons as a proxy for neuronal activity in multiple animal models, including Caenorhabditis elegans, zebrafish, Drosophila, and mice.[19] Recently, genetically encoded voltage indicators (GEVIs) have been developed alongside GECIs to more directly probe neuronal activity at the cellular level in these animal models.[20]
GCaMP has played a vital role in establishing large-scale neural recordings in animals to investigate how activity patterns in neuronal networks influence behavior. For example, Nguyen et al. (2016) used GCaMP in whole-brain imaging during free movement of C. elegans to identify neurons and groups of neurons whose activity correlated with specific locomotor behaviors.[21]
Muto et al. (2003) expressed GCaMP in zebrafish embryos to measure and map the coordinated activity of spinal motor neurons to different parts of the brain during the onset, propagation, and recovery of seizures induced by pentylenetetrazol.[22] GCaMP expression in zebrafish brains has also been used to study activation of neural circuits in cognitive processes like prey capture, impulse control, and attention.[23][24]
Additionally, researchers have used GCaMP to observe neuronal activity in mice by expressing it under control of the Thy1 promoter, which is found in excitatory pyramidal neurons.[25] For instance, integration of neurons into circuits during motor learning has been tracked by using GCaMP to observe synchronized fluctuation patterns in Ca2+ levels.[26][27][28] GCaMP has also been used to observe Ca2+ dynamics in subcellular compartments of mouse neurons: Cichon and Gan (2015) used GCaMP to show that neurons in the mouse motor cortex exhibit NMDA-driven increases in Ca2+ that are independent for each dendritic spine, thus showing that individual dendritic spines regulate synaptic plasticity.[29] Finally, GCaMP has been used to identify activity patterns in specific regions of the mouse brain. For instance, Jones et al. (2018) used GCaMP6 in mice to measure neuronal activity in the suprachiasmatic nucleus (SCN), the mammalian circadian pacemaker, and showed that SCN neurons that produced vasoactive intestinal peptide (VIP) exhibited daily activity rhythms in vivo that correlated with VIP release.[30]
GCaMP has also been combined with fiber photometry to measure population-level Ca2+ changes within subpopulations of neurons in freely moving animals.[31] For instance, Clarkson et al. (2017) used this method to show that neurons in the arcuate nucleus of the hypothalamus synchronize to increases in Ca2+ immediately prior to pulses of luteinizing hormone (LH).[32] While GCaMP imaging with fiber photometry cannot track changes in Ca2+ levels within individual neurons, it provides greater temporal resolution for large-scale changes.[33]
Cardiac conduction
Ca2+ currents through cardiomyocyte gap junctions mediate synchronized contraction of cardiac tissue. As a result, GCaMP expression in cardiomyocytes, both in vitro and in vivo, has been used to study Ca2+-influx-dependent excitation and contraction in zebrafish and mice.[34] For instance, Tallini et al. (2006) expressed GCaMP2 in mouse embryos to show that, at embryonic day 10.5, electrical conduction was rapid in the atria and ventricles but slow in the atrioventricular canal.[8] Chi et al. (2008) used a transgenic cardiac-specific GCaMP zebrafish line to image cardiomyocyte activation throughout the cardiac cycle; from their results, they characterized four developmental stages of the zebrafish cardiac conduction system and identified 17 novel mutations affecting cardiac conduction.[35] However, uncontrolled expression of GCaMP leads to cardiac hypertrophy due to overexpression of the calmodulin motif, which interferes with intracellular calcium signaling. As a result, experiments using cardiac tissue should carefully control the level of GCaMP expression.[8]
Signaling pathway activation
Since Ca2+ is a common second messenger, GCaMP has been used to monitor the activation of signaling pathways. For instance, Bonder and McCarthy (2014) used GCaMP to show that astrocytic G-protein coupled receptor (GPCR) signaling and subsequent Ca2+ release was not responsible for neurovascular coupling, the process by which changes in neuronal activity lead to changes in local blood flow.[36] Similarly, Greer and Bear et al. (2016) used GCaMP to characterize the dynamics of Ca2+ influx in necklace olfactory neuron signaling, which uses transmembrane MS4A proteins as chemoreceptors.[37]
See also
References
- 1 2 3 Nakai J, Ohkura M, Imoto K (February 2001). "A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein". Nature Biotechnology. 19 (2): 137–41. doi:10.1038/84397. PMID 11175727. S2CID 30254550.
- 1 2 3 Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. (July 2013). "Ultrasensitive fluorescent proteins for imaging neuronal activity". Nature. 499 (7458): 295–300. Bibcode:2013Natur.499..295C. doi:10.1038/nature12354. PMC 3777791. PMID 23868258.
- ↑ Barnett LM, Hughes TE, Drobizhev M (2017-02-09). "Deciphering the molecular mechanism responsible for GCaMP6m's Ca2+-dependent change in fluorescence". PLOS ONE. 12 (2): e0170934. Bibcode:2017PLoSO..1270934B. doi:10.1371/journal.pone.0170934. PMC 5300113. PMID 28182677.
- ↑ Whitaker M (2010-01-01). "Genetically encoded probes for measurement of intracellular calcium". Calcium in Living Cells. Methods in Cell Biology. Vol. 99. pp. 153–82. doi:10.1016/B978-0-12-374841-6.00006-2. ISBN 9780123748416. PMC 3292878. PMID 21035686.
- 1 2 Ji G, Feldman ME, Deng KY, Greene KS, Wilson J, Lee JC, et al. (May 2004). "Ca2+-sensing transgenic mice: postsynaptic signaling in smooth muscle". The Journal of Biological Chemistry. 279 (20): 21461–8. doi:10.1074/jbc.M401084200. PMID 14990564. S2CID 9532711.
- ↑ Akerboom J, Rivera JD, Guilbe MM, Malavé EC, Hernandez HH, Tian L, et al. (March 2009). "Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design". The Journal of Biological Chemistry. 284 (10): 6455–64. doi:10.1074/jbc.M807657200. PMC 2649101. PMID 19098007.
- ↑ Wang Q, Shui B, Kotlikoff MI, Sondermann H (December 2008). "Structural basis for calcium sensing by GCaMP2". Structure. 16 (12): 1817–27. doi:10.1016/j.str.2008.10.008. PMC 2614139. PMID 19081058.
- 1 2 3 4 Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, et al. (March 2006). "Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2". Proceedings of the National Academy of Sciences of the United States of America. 103 (12): 4753–8. Bibcode:2006PNAS..103.4753T. doi:10.1073/pnas.0509378103. PMC 1450242. PMID 16537386.
- ↑ Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, et al. (December 2009). "Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators". Nature Methods. 6 (12): 875–81. doi:10.1038/nmeth.1398. PMC 2858873. PMID 19898485.
- ↑ Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, et al. (October 2012). "Optimization of a GCaMP calcium indicator for neural activity imaging". The Journal of Neuroscience. 32 (40): 13819–40. doi:10.1523/JNEUROSCI.2601-12.2012. PMC 3482105. PMID 23035093.
- 1 2 Inoue M (June 2020). "Genetically encoded calcium indicators to probe complex brain circuit dynamics in vivo". Neuroscience Research. 169: 2–8. doi:10.1016/j.neures.2020.05.013. PMID 32531233. S2CID 219559849.
- 1 2 3 4 Haery L. "Switch to GECO? An Overview of AAV Encoded Calcium Sensors". blog.addgene.org. Retrieved 2021-05-06.
- ↑ Bassett JJ, Monteith GR (2017-01-01). "Genetically Encoded Calcium Indicators as Probes to Assess the Role of Calcium Channels in Disease and for High-Throughput Drug Discovery". Ion Channels Down Under (PDF). Advances in Pharmacology. Vol. 79. pp. 141–171. doi:10.1016/bs.apha.2017.01.001. ISBN 9780128104132. PMID 28528667.
- ↑ Yang Y, Liu N, He Y, Liu Y, Ge L, Zou L, et al. (April 2018). "Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP". Nature Communications. 9 (1): 1504. Bibcode:2018NatCo...9.1504Y. doi:10.1038/s41467-018-03719-6. PMC 5904127. PMID 29666364.
- ↑ Zhang Y, Rózsa M, Bushey D, Reep D, Broussard GY, Tsang A, et al. (2020). "jGCaMP8 Fast Genetically Encoded Calcium Indicators": 361685 Bytes. doi:10.25378/janelia.13148243.v4.
{{cite journal}}: Cite journal requires|journal=(help) - 1 2 3 4 Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, et al. (March 2016). Häusser M (ed.). "Sensitive red protein calcium indicators for imaging neural activity". eLife. 5: e12727. doi:10.7554/eLife.12727. PMC 4846379. PMID 27011354.
- 1 2 Akerboom J, Carreras Calderón N, Tian L, Wabnig S, Prigge M, Tolö J, et al. (2013). "Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics". Frontiers in Molecular Neuroscience. 6: 2. doi:10.3389/fnmol.2013.00002. PMC 3586699. PMID 23459413.
- ↑ Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, et al. (September 2011). "An expanded palette of genetically encoded Ca²⁺ indicators". Science. 333 (6051): 1888–91. doi:10.1126/science.1208592. PMC 3560286. PMID 21903779.
- ↑ Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, et al. (July 2019). "High-performance calcium sensors for imaging activity in neuronal populations and microcompartments". Nature Methods. 16 (7): 649–657. doi:10.1038/s41592-019-0435-6. PMID 31209382. S2CID 189927684.
- ↑ Yang, Helen H.; St-Pierre, François (2016-09-28). "Genetically Encoded Voltage Indicators: Opportunities and Challenges". Journal of Neuroscience. 36 (39): 9977–9989. doi:10.1523/JNEUROSCI.1095-16.2016. ISSN 0270-6474. PMC 5039263. PMID 27683896.
- ↑ Nguyen JP, Shipley FB, Linder AN, Plummer GS, Liu M, Setru SU, et al. (February 2016). "Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans". Proceedings of the National Academy of Sciences of the United States of America. 113 (8): E1074-81. arXiv:1501.03463. Bibcode:2016PNAS..113E1074N. doi:10.1073/pnas.1507110112. PMC 4776509. PMID 26712014.
- ↑ Muto A, Ohkura M, Kotani T, Higashijima S, Nakai J, Kawakami K (March 2011). "Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish". Proceedings of the National Academy of Sciences of the United States of America. 108 (13): 5425–30. Bibcode:2011PNAS..108.5425M. doi:10.1073/pnas.1000887108. PMC 3069178. PMID 21383146.
- ↑ Muto A, Kawakami K (2013). "Prey capture in zebrafish larvae serves as a model to study cognitive functions". Frontiers in Neural Circuits. 7: 110. doi:10.3389/fncir.2013.00110. PMC 3678101. PMID 23781176.
- ↑ Parker MO, Brock AJ, Walton RT, Brennan CH (2013). "The role of zebrafish (Danio rerio) in dissecting the genetics and neural circuits of executive function". Frontiers in Neural Circuits. 7: 63. doi:10.3389/fncir.2013.00063. PMC 3619107. PMID 23580329.
- ↑ Chen Q, Cichon J, Wang W, Qiu L, Lee SJ, Campbell NR, et al. (October 2012). "Imaging neural activity using Thy1-GCaMP transgenic mice". Neuron. 76 (2): 297–308. doi:10.1016/j.neuron.2012.07.011. PMC 4059513. PMID 23083733.
- ↑ Peters AJ, Chen SX, Komiyama T (June 2014). "Emergence of reproducible spatiotemporal activity during motor learning". Nature. 510 (7504): 263–7. Bibcode:2014Natur.510..263P. doi:10.1038/nature13235. PMID 24805237. S2CID 4463927.
- ↑ Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, et al. (March 2013). "Long-term dynamics of CA1 hippocampal place codes". Nature Neuroscience. 16 (3): 264–6. doi:10.1038/nn.3329. PMC 3784308. PMID 23396101.
- ↑ Lin MZ, Schnitzer MJ (August 2016). "Genetically encoded indicators of neuronal activity". Nature Neuroscience. 19 (9): 1142–53. doi:10.1038/nn.4359. PMC 5557009. PMID 27571193.
- ↑ Cichon J, Gan WB (April 2015). "Branch-specific dendritic Ca(2+) spikes cause persistent synaptic plasticity". Nature. 520 (7546): 180–5. Bibcode:2015Natur.520..180C. doi:10.1038/nature14251. PMC 4476301. PMID 25822789.
- ↑ Jones JR, Simon T, Lones L, Herzog ED (September 2018). "SCN VIP Neurons Are Essential for Normal Light-Mediated Resetting of the Circadian System". The Journal of Neuroscience. 38 (37): 7986–7995. doi:10.1523/JNEUROSCI.1322-18.2018. PMC 6596148. PMID 30082421.
- ↑ Han SY, Clarkson J, Piet R, Herbison AE (November 2018). "Optical Approaches for Interrogating Neural Circuits Controlling Hormone Secretion". Endocrinology. 159 (11): 3822–3833. doi:10.1210/en.2018-00594. PMID 30304401. S2CID 52954832.
- ↑ Clarkson J, Han SY, Piet R, McLennan T, Kane GM, Ng J, et al. (November 2017). "Definition of the hypothalamic GnRH pulse generator in mice". Proceedings of the National Academy of Sciences of the United States of America. 114 (47): E10216–E10223. Bibcode:2017PNAS..11410216C. doi:10.1073/pnas.1713897114. PMC 5703322. PMID 29109258.
- ↑ Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, et al. (June 2014). "Natural neural projection dynamics underlying social behavior". Cell. 157 (7): 1535–51. doi:10.1016/j.cell.2014.05.017. PMC 4123133. PMID 24949967.
- ↑ Kaestner L, Scholz A, Tian Q, Ruppenthal S, Tabellion W, Wiesen K, et al. (May 2014). "Genetically encoded Ca2+ indicators in cardiac myocytes". Circulation Research. 114 (10): 1623–39. doi:10.1161/CIRCRESAHA.114.303475. PMID 24812351. S2CID 10856784.
- ↑ Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, et al. (May 2008). "Genetic and physiologic dissection of the vertebrate cardiac conduction system". PLOS Biology. 6 (5): e109. doi:10.1371/journal.pbio.0060109. PMC 2430899. PMID 18479184.
- ↑ Bonder DE, McCarthy KD (September 2014). "Astrocytic Gq-GPCR-linked IP3R-dependent Ca2+ signaling does not mediate neurovascular coupling in mouse visual cortex in vivo". The Journal of Neuroscience. 34 (39): 13139–50. doi:10.1523/JNEUROSCI.2591-14.2014. PMC 4172806. PMID 25253859.
- ↑ Greer PL, Bear DM, Lassance JM, Bloom ML, Tsukahara T, Pashkovski SL, et al. (June 2016). "A Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction". Cell. 165 (7): 1734–1748. doi:10.1016/j.cell.2016.05.001. PMC 4912422. PMID 27238024.