Graphical time warping (GTW) is a framework for jointly aligning multiple pairs of time series or sequences.[1] GTW considers both the alignment accuracy of each sequence pair and the similarity among pairs. On contrary, alignment with dynamic time warping (DTW) considers the pairs independently and minimizes only the distance between the two sequences in a given pair. Therefore, GTW generalizes DTW and could achieve a better alignment performance when similarity among pairs is expected.
One application of GTW is signal propagation analysis in time-lapse bio-imaging data, where the propagation patterns in adjacent pixels are generally similar. Other applications include signature identification, binocular stereo depth calculation, and liquid chromatography–mass spectrometry (LC-MS) profile alignment in proteomics data analysis.[2] Indeed, as long as the data are structured with inter-dependent time series/sequences, they can be analyzed with GTW.
GTW is able to model constraints or similarities between warping paths by transforming the DTW-equivalent shortest path problem to the maximum flow problem in the dual graph, which can be solved by most max-flow algorithms. However, when the data is large, these algorithms become time-consuming and the memory usage is high. An efficient algorithm, Bidirectional pushing with Linear Component Operations (BILCO),[3] was developed to solve the GTW problem. It could achieve an average 10-fold improvement in both computational and memory usage compared with the state of art generic maximum flow algorithms in GTW applications.
Joint alignment and GTW formulation
Joint alignment
Assume there are pairs of time series , and each pair has a corresponding warping path . Some pairs of warping paths are known to be similar, and the set of all such pairs is denoted as . For example, if is in this set, warping paths and are similar. To optimize both the similarity between the aligned time series and the warping paths distances, the joint alignment problem is formulated as a minimization problem:
Here denotes the distance between and after alignment with warping function , is the distance between warping paths and defined by the area of the region bounded by and , and is a hyperparameter balancing the time series alignment cost term and warping function distance term.
Notice that the similarity strength can be application-specific or user-designed. For different related warping paths pair , we can set different parameters . For simplicity, here we use the unified hyperparameter .
The above minimization problem is intuitively formulated. However, it is not clear how to efficiently solve it in its original form, and a naïve enumeration of the warping paths leads to an NP-hard problem.
GTW formulation

This minimization problem can be reformulated into a minimum cut problem on a special graph termed GTW graph, where the minimum cut and the warping paths are equivalent.[1] The formulation could be described as:
- For the time-series pair, construct its DTW graph. Then convert this DTW graph to its dual graph, termed as GTW subgraph . Set the capacities of reverse edges to be infinite.
- For each pair of similar warping paths and , linking the nodes of the same position in and by bidirected edges with capacity . Such edges are termed as cross edges.
- The constructed GTW graph, as shown in the figure, consists of GTW subgraphs and cross edges.
- Using maximum flow algorithms to obtain the minimum cut of the constructed graph. The minimum cut within each GTW subgraph corresponds to one warping path.
Explanation of the equivalence
Each GTW subgraph is the dual graph of the DTW graph representing the alignment of a single time-series pair. As a result, the cut within a GTW subgraph is dual to a warping path in DTW graph, and the profile alignment cost term can be represented by the cut cost within subgraphs. The infinite capacities of reverse edges are used to guarantee the monotonicity and continuity of warping paths.
Cross edges constrain the similarity of warping paths and contribute to the distance term in the objective function. Notice that in a minimum cut problem, the nodes would eventually be assigned to the source side or the sink side, and the final cut is defined by the edges between two sides. Each pair of mismatched nodes in and contribute to the distance between and and would result in an extra cost. Thus, the distance term could be represented by the cut cost in cross edges.
Therefore, the cut cost in the GTW graph corresponds to the cost terms in the objective function. Recalling the fact that the cut within each subgraph corresponds to the warping path of one time-series pair, the minimum cut of GTW graph corresponds to the optimal solution of warping paths in the joint alignment.
Extension
Neighbor-wise Compound-specific Graphical Time Warping (ncGTW)
In multiple sequence alignment, the purpose is to align all sequences to a common reference. However, this common reference is usually unknown. In addition, there is also structural information among the sequences. Though GTW cannot be directly applied in these applications, a two-stage framework called ncGTW was built upon GTW to solve this problem. In the first stage, the prior structural knowledge among the sequences is utilized to obtain the warping functions. In the second stage, these warping functions help to jointly align all sequences to a virtual reference, which does not need to be explicitly specified. ncGTW was applied to LC-MS profile alignment problems in proteomics data and performed better than existing approaches.[2]
Efficient algorithm
Bidirectional pushing with Linear Component Operations (BILCO)
Solving the minimum cut problem on GTW graph through traditional maximum flow algorithms would take a long running time and large memory usage due to the large graph size, which limits the usage of GTW. BILCO algorithm utilizes two important properties of the joint alignment problem and achieves an average 10-fold improvement in both running time and memory usage. The two properties are:
- Joint alignment problem is a generalization of pairwise alignment and there are numerous DTW problems embedded in GTW graph. As each GTW subgraph is dual to a DTW graph, the maximum flow within each GTW subgraph can be solved in linear time through dynamic programming.
- In many applications, a rough approximate solution of the warping paths can be estimated, which could serve as the initialization to accelerate the solving process.
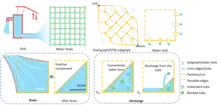
According to the first property, BILCO divides the flow exchange into two types: (1) Flow exchange within GTW subgraph; (2) Flow exchange across related GTW subgraphs. The process can be analogized to the process of pumping water from connected water tanks, and two types of flow exchange are termed Drain and Discharge. To fully utilize such property, components (each component is a connected subset of GTW subgraph), rather than single nodes, are used as the operation unit. Both Drain and Discharge component operations can be implemented in linear time.
The second property inspires the bidirectional-pushing strategy. In this strategy, BILCO first segments the graph into two parts using the initial approximate solution, and then pushes excess/deficit in obtained sink/source parts, respectively. Compared with existing push-relabel-based maximum flow algorithms, BILCO significantly reduces redundant computation. It is worth noting that such a strategy could be also utilized to help accelerate other push-relabel-based algorithms.[3]
Applications
Signal propagation analysis
In time-lapse bio-imaging data, signal propagation is a widely observed phenomenon in many cell types.[4] Studying signal propagation may help uncover the function of these cells in both normal and pathological conditions. The propagation information could be derived from the warping paths by aligning pixels’ curves with a reference signal. Due to the low signal-to-noise ratio in bio-imaging data, pairwise alignment methods usually lead to unsatisfactory results. Considering the spatial correlation of the signals, the similarity of warping paths between adjacent pixels can be utilized in GTW to enhance the alignment performance, which may lead to a more accurate calculation of propagation properties.
Depth extraction
In binocular stereo images, alignment technique can be used to extract depth information.[5] The depth could be derived by the disparity of the same row between left image and right image. Since the depth of adjacent rows should be similar, GTW could be utilized to enhance the extraction result.
Signature identification
A signature usually contains multiple feature sequences, such as the x location, the y location, and the pressure.[6] Those feature sequences are correlated, which indicates that when comparing two signatures, the distance measure obtained by pairwise alignment is not optimal. GTW could take the dependency between features into account and provide a better distance measure.
Biological sequence alignment
In a biological sequence data set, it is common that there is some structural information among the sequences. In LC-MS data, the samples of nearby profiles tend to have similar patterns of distortion and GTW is extended to jointly align these profiles. The same technique may also be applied to the joint alignment of other sequences. Structural information between sequences also exists in DNA and amino acids data. For example, the sequences between related species are more similar compared with sequences from more remotely related species. This information could be utilized by GTW.
See also
References
- 1 2 Wang, Yizhi; Miller, David J; Poskanzer, Kira; Wang, Yue; Tian, Lin; Yu, Guoqiang (2016). "Graphical Time Warping for Joint Alignment of Multiple Curves". Advances in Neural Information Processing Systems. Curran Associates, Inc. 29.
- 1 2 Wu, Chiung-Ting; Wang, Yizhi; Wang, Yinxue; Ebbels, Timothy; Karaman, Ibrahim; Graça, Gonçalo; Pinto, Rui; Herrington, David M; Wang, Yue; Yu, Guoqiang (1 May 2020). "Targeted realignment of LC-MS profiles by neighbor-wise compound-specific graphical time warping with misalignment detection". Bioinformatics. 36 (9): 2862–2871. doi:10.1093/bioinformatics/btaa037. PMC 7203744. PMID 31950989.
- 1 2 Mi, Xuelong; Wang, Mengfan; Chen, Alex; Lim, Jing-Xuan; Wang, Yizhi; Ahrens, Misha B.; Yu, Guoqiang (6 December 2022). "BILCO: An Efficient Algorithm for Joint Alignment of Time Series". Advances in Neural Information Processing Systems. 35: 36270–36281.
- ↑ Wang, Yizhi; DelRosso, Nicole V.; Vaidyanathan, Trisha V.; Cahill, Michelle K.; Reitman, Michael E.; Pittolo, Silvia; Mi, Xuelong; Yu, Guoqiang; Poskanzer, Kira E. (November 2019). "Accurate quantification of astrocyte and neurotransmitter fluorescence dynamics for single-cell and population-level physiology". Nature Neuroscience. 22 (11): 1936–1944. doi:10.1038/s41593-019-0492-2. ISSN 1546-1726. PMC 6858541. PMID 31570865.
- ↑ Ishikawa, Hiroshi; Geiger, Davi (1998). "Occlusions, discontinuities, and epipolar lines in stereo". Computer Vision — ECCV'98. Lecture Notes in Computer Science. Vol. 1406. Springer. pp. 232–248. doi:10.1007/BFb0055670. ISBN 978-3-540-64569-6.
- ↑ Okawa, Manabu (2019). "Template Matching Using Time-Series Averaging and DTW With Dependent Warping for Online Signature Verification". IEEE Access. 7: 81010–81019. doi:10.1109/ACCESS.2019.2923093. S2CID 195774867.