Lysenin is a pore-forming toxin (PFT) present in the coelomic fluid of the earthworm Eisenia fetida. Pore-forming toxins are a group of proteins that act as virulence factors of several pathogenic bacteria. Lysenin proteins are chiefly involved in the defense against cellular pathogens.[1] Following the general mechanism of action of PFTs lysenin is segregated as a soluble monomer that binds specifically to a membrane receptor, sphingomyelin in the case of lysenin. After attaching to the membrane, the oligomerization begins, resulting in a nonamer on top of membrane, known as a prepore. After a conformational change, which could be triggered by a decrease of pH, the oligomer is inserted into the membrane in the so-called pore state.
Monomer
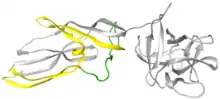
Lysenin is a protein produced in the coelomocyte-leucocytes of the earthworm Eisenia fetida.[2] This protein was first isolated from the coelomic fluid in 1996 and named lysenin (from lysis and Eisenia).[3] Lysenin is a relatively small water-soluble molecule with a molecular weight of 33 kDa. Using X-ray crystallography, lysenin was classified as a member of the Aerolysin protein family by structure and function.[4] Structurally, each lysenin monomer consists of a receptor binding domain (grey globular part on right of Figure 1) and a Pore Forming Module (PFM); domains shared throughout the aerolysin family.[4] The lysenin receptor binding domain shows three sphingomyelin binding motifs. The Pore Forming Module contains the regions that undergo large conformational changes to become the β-barrel in the pore.[5]
Membrane receptors
The natural membrane target of lysenin is an animal plasma membrane lipid called sphingomyelin located mainly in its outer leaflet, involving at least three of its phosphatidylcholines (PC) groups.[6] Sphingomyelin is usually found associated with cholesterol in lipid rafts.[7] Cholesterol, which enhances oligomerization, provides a stable platform with high lateral mobility where monomer-monomer encounters are more probable.[6] PFTs have shown to be able to remodel the membrane structure,[8] sometimes even mixing lipid phases.[9]
The region of the lysenin pore β-barrel expected to be immersed in the hydrophobic region of the membrane is the 'detergent belt', the 3.2 nm high region occupied by detergent in Cryogenic Electron Microscopy (Cryo-EM) studies of the pore.[10] On the other hand, sphingomyelin/Cholesterol bilayers are about 4.5 nm height.[11] This difference in height between the detergent belt and the sphingomyelin/cholesterol bilayer implies a bend of the membrane in the region surrounding the pore, called negative mismatch.[12] This bending results in a net attraction between pores that induce pores aggregation.
Binding, oligomerization and insertion
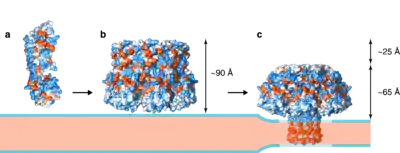
Membrane binding is a requisite to initiate PFT oligomerization. Lysenin monomers bind specifically to sphingomyelin via the receptor binding domain.[13] The final lysenin oligomer is constituted by nine monomers without quantified deviations.[14] When lysenin monomers bind to sphingomyelin-enriched membrane regions, they provide a stable platform with a high lateral mobility, hence favouring the oligomerization.[15] As with most PFTs, lysenin oligomerization occurs in a two-step process, as was recently imaged.
The process begins with monomers being adsorbed into the membrane by specific interactions, resulting in an increased concentration of monomers. This increase is promoted by the small area where the membrane receptor accumulates owing to the fact that the majority of PFT membrane receptors are associated with lipid rafts.[16] Another side effect, aside from the increase of monomer concentration, is the monomer-monomer interaction. This interaction increases lysenin oligomerization. After a critical threshold concentration is reached, several oligomers are formed simultaneously, although sometimes these are incomplete.[17] In contrast to PFTs of the cholesterol-dependent cytolysin family,[18] the transition from incomplete lysenin oligomers to complete oligomers has not been observed.
A complete oligomerization results in the so-called prepore state, a structure on the membrane. Determining the prepore's structure by X-ray or Cryo-EM is a challenging process that so far has not produced any results. The only available information about the prepore structure was provided by Atomic Force Microscopy (AFM). The measured prepore height was 90 Å; and the width 118 Å, with an inner pore of 50 Å.[17] A model of the prepore was built aligning the monomer structure (PDB: 3ZXD) with the pore structure (PDB: 5GAQ) by their receptor-binding domains (residues 160 to 297). A recent study in aerolysin suggests that the currently accepted model for the lysenin prepore should be revisited, according to the new available data on the aerolysin insertion.[19]
A conformational change transforms the PFM into the transmembrane β-barrel, leading to the pore state.[20] The trigger mechanism for the prepore-to-pore transition in lysenin depends on three glutamic acid residues (E92, E94 and E97), and is activated by a decrease in pH,[21] from physiological conditions to the acidic conditions reached after endocytosis, or an increase in calcium extracellular concentration.[22] These three glutamic acids are located in an α-helix that forms part of the PFM, and glutamic acids are found in aerolysin family members in its PFMs. Such a conformational change produces a decrease in the oligomer height of 2.5 nm according to AFM measurements.[17] The main dimensions, using lysenin pore X-ray structure, are height 97 Å, width 115 Å and the inner pore of 30 Å.[20] However, complete oligomerization into the nonamer is not a requisite for the insertion, since incomplete oligomers in the pore state can be found.[17] The prepore to pore transition can be blocked in crowded conditions, a mechanism that could be general to all β-PFTs. The first hint of crowding effect on prepore to pore transition was given by congestion effects in electrophysiology experiments.[23] High-Speed AFM studies incubating lysenin on sphingomyelin/cholesterol membranes has shown that under crowded conditions the prepore to pore transition gets blocked by steric interactions.[24][25][26]
Insertion consequences
The ultimate consequences of lysenin pore formation are not well documented; however, it is thought to induce apoptosis via three possible hypotheses:
- Breaking the sphingomyelin asymmetry between the two leaflets of the lipid bilayer by punching holes in the membrane[27] and inducing lipid flip-flop (reorientation of a lipid from one leaflet of a membrane bilayer to the other).[28]
- Increasing the calcium concentration in the cytoplasm.[29]
- Decreasing the potassium concentration in the cytoplasm.[30]
Biological role
The biological role of lysenin remains unknown. It has been suggested that lysenin may play a role as a defence mechanism against attackers such as bacteria, fungi or small invertebrates.[31] However, lysenin's activity is dependent upon binding to sphingomyelin, which is not present in the membranes of bacteria, fungi or most invertebrates. Rather, sphingomyelin is mainly present in the plasma membrane of chordates.[32] Another hypothesis is that the earthworm, which is able to expel coelomic fluid under stress,[33][34] generates an avoidance behaviour to its vertebrate predators (such as birds, hedgehogs or moles).[35] If that is the case, the expelled lysenin might be more effective if the coelomic fluid reaches the eye, where the concentration of sphingomyelin is ten times higher than in other body organs.[36] A complementary hypothesis is that the pungent smell of the coelomic fluid - giving the earthworm its specific epithet foetida - is an anti-predator adaptation. However, it remains unknown whether lysenin contributes to avoidance of Eisenia by predators.[37]
Applications
Lysenin's conductive properties have been studied for years.[38] Like most pore-forming toxins, lysenin forms a non-specific channel that is permeable to ions, small molecules, and small peptides.[39] There have also been over three decades of studies into finding suitable pores for converting into nanopore sequencing systems that can have their conductive properties tuned by point mutation.[40] Owing to its binding affinity for sphingomyelin, lysenin (or just the receptor binding domain) has been used as a fluorescence marker to detect the sphingomyelin domain in membranes.[41]
References
![]() This article was submitted to WikiJournal of Science for external academic peer review in 2019 (reviewer reports). The updated content was reintegrated into the Wikipedia page under a CC-BY-SA-3.0 license (2019). The version of record as reviewed is:
Ignacio L. B. Munguira; et al. (17 August 2019). "Lysenin" (PDF). WikiJournal of Science. 2 (1): 6. doi:10.15347/WJS/2019.006. ISSN 2470-6345. Wikidata Q76846397.
This article was submitted to WikiJournal of Science for external academic peer review in 2019 (reviewer reports). The updated content was reintegrated into the Wikipedia page under a CC-BY-SA-3.0 license (2019). The version of record as reviewed is:
Ignacio L. B. Munguira; et al. (17 August 2019). "Lysenin" (PDF). WikiJournal of Science. 2 (1): 6. doi:10.15347/WJS/2019.006. ISSN 2470-6345. Wikidata Q76846397.
- ↑ Bruhn, Heike; Winkelmann, Julia; Andersen, Christian; Andrä, Jörg; Leippe, Matthias (2006). "Dissection of the mechanisms of cytolytic and antibacterial activity of lysenin, a defence protein of the annelid Eisenia fetida". Developmental and Comparative Immunology. 30 (7): 597–606. doi:10.1016/j.dci.2005.09.002. PMID 16386304.
- ↑ Yilmaz, N.; Yamaji-Hasegawa, A.; Hullin-Matsuda, F.; Kobayashi, T. (2018). "Molecular mechanisms of action of sphingomyelin-specific pore-forming toxin, lysenin". Seminars in Cell & Developmental Biology. 73: 188–198. doi:10.1016/j.semcdb.2017.07.036. PMID 28751253.
- ↑ Sekizawa, Y.; Hagiwara, K.; Nakajima, T.; Kobayashi, H. (1996). "A Novel Protein, Lysenin, that Causes Contraction of the Isolate Rat Aorta: Its Purification from the Coelomic Fluid of the Earthworm, Eisenia foetida". Biomedical Research. 17 (3): 197–203. doi:10.2220/biomedres.17.197.
- 1 2 De Colibus, L.; Sonnen, A. F.-P.; Morris, K. J.; Siebert, C. A.; Abrusci, P.; Plitzko, J.; Hodnik, V.; Leippe, M.; Volpi, E.; Anderluh, G.; Gilbert, R. J. C. (2012). "Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins And Its Mode of Sphingomyelin Recognition". Structure. 20 (9): 1498–1507. doi:10.1016/j.str.2012.06.011. PMC 3526787. PMID 22819216.
- ↑ Bokori-Brown, M.; Martin, T. G.; Naylor, C. E.; Basak, A. K.; Titball, R. W.; Savva, C. G. (2016). "Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein". Nature Communications. 7 (1): 11293. Bibcode:2016NatCo...711293B. doi:10.1038/ncomms11293. PMC 4823867. PMID 27048994.
- 1 2 Ishitsuka, R.; Kobayashi, T. (2007). "Cholesterol and Lipid/Protein Ratio Control the Oligomerization of a Sphingomyelin-Specific Toxin, Lysenin". Biochemistry. 46 (6): 1495–1502. doi:10.1021/bi061290k. PMID 17243772. S2CID 22016219.
- ↑ Simons, K.; Gerl, M. J. (2010). "Revitalizing membrane rafts: new tools and insights". Nature Reviews Molecular Cell Biology. 11 (10): 688–699. doi:10.1038/nrm2977. PMID 20861879. S2CID 1866391.
- ↑ Ros, U.; García-Sáez, A. J. (2015). "More Than a Pore: The Interplay of Pore-Forming Proteins and Lipid Membranes". The Journal of Membrane Biology. 248 (3): 545–561. doi:10.1007/s00232-015-9820-y. PMID 26087906. S2CID 16305100.
- ↑ Yilmaz, N.; Kobayashi, T. (2015). "Visualization of Lipid Membrane Reorganization Induced by a Pore-Forming Toxin Using High-Speed Atomic Force Microscopy". ACS Nano. 9 (8): 7960–7967. doi:10.1021/acsnano.5b01041. PMID 26222645.
- ↑ Bokori-Brown, M.; Martin, T. G.; Naylor, C. E.; Basak, A. K.; Titball, R. W.; Savva, C. G. (2016). "Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein". Nature Communications. 7 (1): 11293. Bibcode:2016NatCo...711293B. doi:10.1038/ncomms11293. PMC 4823867. PMID 27048994.
- ↑ Quinn, P. J. (2013). "Structure of Sphingomyelin Bilayers and Complexes with Cholesterol Forming Membrane Rafts". Langmuir. 29 (30): 9447–9456. doi:10.1021/la4018129. PMID 23863113.
- ↑ Guigas, G.; Weiss, M. (2016). "Effects of protein crowding on membrane systems". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1858 (10): 2441–2450. doi:10.1016/j.bbamem.2015.12.021. PMID 26724385.
- ↑ De Colibus, L.; Sonnen, A. F.-P.; Morris, K. J.; Siebert, C. A.; Abrusci, P.; Plitzko, J.; Hodnik, V.; Leippe, M.; Volpi, E.; Anderluh, G.; Gilbert, R. J. C. (2012). "Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins And Its Mode of Sphingomyelin Recognition". Structure. 20 (9): 1498–1507. doi:10.1016/j.str.2012.06.011. PMC 3526787. PMID 22819216.
- ↑ Munguira, I.; Casuso, I.; Takahashi, H.; Rico, F.; Miyagi, A.; Chami, M.; Scheuring, S. (2016). "Glasslike Membrane Protein Diffusion in a Crowded Membrane" (PDF). ACS Nano. 10 (2): 2584–2590. doi:10.1021/acsnano.5b07595. PMID 26859708. S2CID 206699095.
- ↑ Ishitsuka, R.; Kobayashi, T. (2007). "Cholesterol and Lipid/Protein Ratio Control the Oligomerization of a Sphingomyelin-Specific Toxin, Lysenin". Biochemistry. 46 (6): 1495–1502. doi:10.1021/bi061290k. PMID 17243772. S2CID 22016219.
- ↑ Lafont, F.; Van Der Goot, F. G. (2005). "Bacterial invasion via lipid rafts". Cellular Microbiology. 7 (5): 613–620. doi:10.1111/j.1462-5822.2005.00515.x. PMID 15839890. S2CID 26547616.
- 1 2 3 4 Yilmaz, N.; Yamada, T.; Greimel, P.; Uchihashi, T.; Ando, T.; Kobayashi, T. (2013). "Real-Time Visualization of Assembling of a Sphingomyelin-Specific Toxin on Planar Lipid Membranes". Biophysical Journal. 105 (6): 1397–1405. Bibcode:2013BpJ...105.1397Y. doi:10.1016/j.bpj.2013.07.052. PMC 3785888. PMID 24047991.
- ↑ Mulvihill, E.; van Pee, K.; Mari, S. A.; Müller, D. J.; Yildiz, Ö. (2015). "Directly Observing the Lipid-Dependent Self-Assembly and Pore-Forming Mechanism of the Cytolytic Toxin Listeriolysin O". Nano Letters. 15 (10): 6965–6973. Bibcode:2015NanoL..15.6965M. doi:10.1021/acs.nanolett.5b02963. PMID 26302195.
- ↑ Iacovache, Ioan; De Carlo, Sacha; Cirauqui, Nuria; Dal Peraro, Matteo; van der Goot, F. Gisou; Zuber, Benoît (2016). "Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process". Nature Communications. 7: 12062. Bibcode:2016NatCo...712062I. doi:10.1038/ncomms12062. PMC 4947156. PMID 27405240.
- 1 2 Bokori-Brown, M.; Martin, T. G.; Naylor, C. E.; Basak, A. K.; Titball, R. W.; Savva, C. G. (2016). "Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein". Nature Communications. 7 (1): 11293. Bibcode:2016NatCo...711293B. doi:10.1038/ncomms11293. PMC 4823867. PMID 27048994.
- ↑ Munguira, I. L. B.; Takahashi, H.; Casuso, I.; Scheuring, S. (2017). "Lysenin Toxin Membrane Insertion Is pH-Dependent but Independent of Neighboring Lysenins". Biophysical Journal. 113 (9): 2029–2036. Bibcode:2017BpJ...113.2029M. doi:10.1016/j.bpj.2017.08.056. PMC 5685674. PMID 29117526.
- ↑ Munguira, I.L.B. (2019). "Lysenin toxin insertion mechanism is Calcium-dependent". bioRxiv. doi:10.1101/771725.
- ↑ Krueger, E.; Bryant, S.; Shrestha, N.; Clark, T.; Hanna, C.; Pink, D.; Fologea, D. (2015). "Intramembrane congestion effects on lysenin channel voltage-induced gating". European Biophysics Journal. 45 (2): 187–194. doi:10.1007/s00249-015-1104-z. PMC 4803513. PMID 26695013.
- ↑ Munguira, I. L. B. (2017). Effect of Crowdedness in the Life Cycle of Lysenin Studied by High-Speed Atomic Force Microscopy (PhD). Aix-Marseille Universite.
- ↑ Munguira, N. L. (2020). "The activity of the pore-forming toxin lysenin is regulated by crowding". Nanotechnology. doi:10.1088/1361-6528/ab5e63. PMID 31796658. S2CID 208620873.
- ↑ Munguira, I. L.B. (2020). "Steric blockage of lysenin toxin by crowding". bioRxiv. doi:10.1101/2020.05.02.073940. S2CID 218539213.
- ↑ Green, D. R. (2000). "Apoptosis and Sphingomyelin Hydrolysis". The Journal of Cell Biology. 150 (1): F5–F8. doi:10.1083/jcb.150.1.F5. PMC 2185551. PMID 10893276.
- ↑ Ros, U.; García-Sáez, A. J. (2015). "More Than a Pore: The Interplay of Pore-Forming Proteins and Lipid Membranes". The Journal of Membrane Biology. 248 (3): 545–561. doi:10.1007/s00232-015-9820-y. PMID 26087906. S2CID 16305100.
- ↑ Orrenius, S.; Zhivotovsky, B.; Nicotera, P. (2003). "Regulation of cell death: the calcium–apoptosis link". Nature Reviews Molecular Cell Biology. 4 (7): 552–565. doi:10.1038/nrm1150. PMID 12838338. S2CID 19079491.
- ↑ Yu, S. P. (2003). "Regulation and critical role of potassium homeostasis in apoptosis". Progress in Neurobiology. 70 (4): 363–386. doi:10.1016/s0301-0082(03)00090-x. PMID 12963093. S2CID 13893235.
- ↑ Ballarin, L.; Cammarata, M. (2016). Lessons in immunity: from single-cell organisms to mammals. Academic Press. ISBN 9780128032527.
- ↑ Kobayashi, H.; Sekizawa, Y.; Aizu, M.; Umeda, M. (2000). "Lethal and non-lethal responses of spermatozoa from a wide variety of vertebrates and invertebrates to lysenin, a protein from the coelomic fluid of the earthworm Eisenia foetida". Journal of Experimental Zoology. 286 (5): 538–549. doi:10.1002/(sici)1097-010x(20000401)286:5<538::aid-jez12>3.0.co;2-w. PMID 10684578.
- ↑ Sukumwang, N.; Umezawa, K. (2013). "Earthworm-Derived Pore-Forming Toxin Lysenin and Screening of Its Inhibitors". Toxins. 5 (8): 1392–1401. doi:10.3390/toxins5081392. PMC 3760042. PMID 23965430.
- ↑ Kobayashi, H.; Ohta, N.; Umeda, M. (2004). "Biology of lysenin, a protein in the coelomic fluid of the earthworm Eisenia foetida". International Review of Cytology. 236: 45–99. doi:10.1016/S0074-7696(04)36002-X. ISBN 9780123646408. PMID 15261736.
- ↑ Swiderska, B.; Kedracka-Krok, S.; Panz, T.; Morgan, A. J.; Falniowski, A.; Grzmil, P.; Plytycz, B. (2017). "Lysenin family proteins in earthworm coelomocytes – Comparative approach". Developmental & Comparative Immunology. 67: 404–412. doi:10.1016/j.dci.2016.08.011. PMID 27567602. S2CID 19895826.
- ↑ Berman, E. R. (1991). Biochemistry of the Eye. Springer. doi:10.1007/978-1-4757-9441-0. ISBN 978-1-4757-9441-0. S2CID 41192657.
- ↑ Edwards, C. A.; Bohlen, P. J. (1996). Biology and Ecology of Earthworms. Springer Science & Business Media. ISBN 978-0-412-56160-3.
- ↑ Bryant, S.; Clark, T.; Thomas, C.; Ware, K.; Bogard, A.; Calzacorta, C.; Prather, D.; Fologea, D. (2018). "Insights into the Voltage Regulation Mechanism of the Pore-Forming Toxin Lysenin". Toxins. 10 (8): 334. doi:10.3390/toxins10080334. PMC 6115918. PMID 30126104.
- ↑ Shrestha, N.; Bryant, S. L.; Thomas, C.; Richtsmeier, D.; Pu, X.; Tinker, J.; Fologea, D. (2017). "Stochastic sensing of Angiotensin II with lysenin channels". Scientific Reports. 7 (1): 2448. Bibcode:2017NatSR...7.2448S. doi:10.1038/s41598-017-02438-0. PMC 5446423. PMID 28550293.
- ↑ Deamer, D.; Akeson, M.; Branton, D. (2016). "Three decades of nanopore sequencing". Nature Biotechnology. 34 (5): 518–524. doi:10.1038/nbt.3423. PMC 6733523. PMID 27153285.
- ↑ Ishitsuka, R.; Kobayashi, T. (2004). "Lysenin: A new tool for investigating membrane lipid organization". Anatomical Science International. 79 (4): 184–190. doi:10.1111/j.1447-073x.2004.00086.x. PMID 15633456. S2CID 1558393.