NAIL-MS (short for nucleic acid isotope labeling coupled mass spectrometry) is a technique based on mass spectrometry used for the investigation of nucleic acids and its modifications. It enables a variety of experiment designs to study the underlying mechanism of RNA biology in vivo. For example, the dynamic behaviour of nucleic acids in living cells, especially of RNA modifications, can be followed in more detail.[1]
Theory
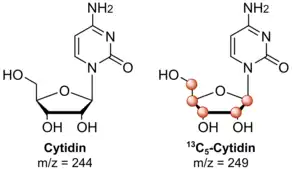
NAIL-MS is used to study RNA modification mechanisms. Therefore, cells in culture are first fed with stable isotope labeled nutrients and the cells incorporate these into their biomolecules. After purification of the nucleic acids, most often RNA, analysis is done by mass spectrometry. Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of ions. Pairs of chemically identical nucleosides of different stable-isotope composition can be differentiated in a mass spectrometer due to their mass difference. Unlabeled nucleosides can therefore be distinguished from their stable isotope labeled isotopologues. For most NAIL-MS approaches it is crucial that the labeled nucleosides are more than 2 Da heavier than the unlabeled ones. This is because 1.1% of naturally occurring carbon atoms are 13C isotopes. In the case of nucleosides this leads to a mass increase of 1 Da in ~10% of the nucleosides. This signal would disturb the final evaluation of the measurement.
NAIL-MS can be used to investigate RNA modification dynamics by changing the labeled nutrients of the corresponding growth medium during the experiment. Furthermore, cell populations can be compared directly with each other without effects of purification bias. Furthermore, it can be used for the production of biosynthetic isotopologues of most nucleosides which are needed for quantification by mass spectrometry and even for the discovery of yet unknown RNA modifications.[2][3][4]
General procedure

In general, cells are cultivated in unlabeled or stable (non-radioactive) isotope labeled media. For example, the medium can contain glucose labeled with six carbon-13 atoms (13C) instead of the normal carbon-12 (12C). Cells growing in this medium, will, depending on model organism, incorporate the heavy glucose into all of their RNA molecules. Thereafter, all nucleotides are 5 Da heavier than their unlabeled isotopologues due to a complete carbon labeling of the ribose. After cultivation and appropriate labeling of the cells, they are generally harvested using phenol/chloroform/guanidinium isothiocyanate. Other extraction methods are possible and sometimes needed (e.g. for yeast). RNA is then isolated by Phenol-Chloroform extraction and iso-Propanol precipitation. Further purification of specific RNA species (e.g. rRNA, tRNA) is usually done by size-exclusion chromatography (SEC) but other approaches are available as well. For most applications the final product needs to be enzymatically digested to nucleosides before analysis by LC-MS. Therefore, digestion enzymes such as benzonase, NP1 and CIP are used.[5][6] Typically, a triple quadrupole in MRM mode is used for the measurements.
Labeling of cells
How the labeling of RNA molecules is achieved depends on the model organism. For E.coli (bacteria) the minimum medium M9 can be used and supplemented with the stable isotope labeled variants of the needed salts. This enables labeling with 13C-carbon, 15N-nitrogen, 34S-sulfur and 2H-hydrogen.[7] In S.cerevisiae (yeast) there are currently two possibilities: First, the use of commercially available complete growth medium, which enables labeling with 13C-carbon and/or 15N-nitrogen and second the use of minimal YNB medium which has to be supplemented with several amino acids and glucose which can be added as stable isotope labeled variants in order to achieve 13C-carbon, 15N-nitrogen and 2H-hydrogen labeling of RNA.[8]
While labeling in model organisms like E.coli and S.cerevisiae is fairly simple, stable isotope labeling in cell culture is much more challenging as the composition of the growth media is much more complex. Neither the supplementation of stable isotope labeled glucose nor the supplementation of stable isotope labeled variants of simple precursors of nucleoside biosynthesis such as glutamine and/or aspartate is sufficient for a defined mass increase higher than 2 Da. Instead, most cells kept in cell culture can be fed with stable isotope labeled methionine for labeling of methyl groups and with stable isotope labeled variants of adenin and uridine for labeling of the nucleoside's base body.[9] Special care must be taken when supplementing the medium with FBS (fetal bovine serum), as it also contains small metabolites used for the biosynthesis of nucleosides. The use of dialyzed FBS is therefore advisable when defined labeling of all nucleosides is desired.
Applications
With NAIL-MS different experiment designs are possible.
Production of SILIS
NAIL-MS can be used to produce stable isotope labeled internal standards (ISTD). Therefore, cells are grown in medium which results in complete labeling of all nucleosides. The purified mix of nucleosides can then be used as ISTD which is needed for accurate absolute quantification of nucleosides by mass spectrometry. This mixture of labeled nucleosides is also referred to as SILIS (stable isotope labeled internal standard).[10] The advantage of this approach is, that all modifications present in an organism can thereby be biosynthesized as labeled compounds. The production of SILIS was already done before the term NAIL-MS emerged.
Comparative Experiments
A comparative NAIL-MS experiment is quite similar to a SILAC experiment but for RNA instead of proteins. First, two populations of the respective cells are cultivated. One of the cell populations is fed with growth medium containing unlabeled nutrients, whereas the second population is fed with growth medium containing stable isotope labeled nutrients. The cells then incorporate the respective isotopologues into their RNA molecules. One of the cell populations serves as a control group whereas the other is subject to the associated research (e.g. KO strain, stress). Upon harvesting of the two cell populations they are mixed and co-processed together to exclude purification-bias. Due to the distinct masses of incorporated nutrients into the nucleosides a differentiation of the two cell populations is possible by mass spectrometry.
Pulse-Chase Experiments
Upon initiation of a pulse-chase experiment the medium is switched from medium(1) to medium(2). The two media must only differ in their isotope content. Thereby it is possible to distinguish between RNA molecules already existent before experiment initiation (= RNA molecules grown in medium(1)) and RNA molecules that are newly transcribed after experiment initiation (= RNA molecules grown in medium(2)). This allows the detailed study of modification dynamics in vivo. The supplementation of labeled methionine in either medium(1) or medium(2) allows the tracing of methylation processes. Other isotopically labeled metabolites potentially allow for further modification analysis.
Altogether NAIL-MS enables the investigation of RNA modification dynamics by mass spectrometry. With this technique, enzymatic demethylation has been observed for several RNA damages inside living bacteria.[4][7]
Discovery of new RNA modifications
For the discovery of uncharacterized modifications cells are grown in unlabeled or 13C‑labeled or 15N‑labeled or 2H‑labeled or 34S‑labeled medium. Unknown signals occurring during mass spectrometry are then inspected in all differentially labeled cultures. If retention times of unknown compounds with appropriately divergent m/z values overlap, a sum formula of the compound can be postulated by calculating the mass differences of the overlapping signal in the differentially labeled cultures. With this method several new RNA modifications could be discovered. This experimental design also was the initial idea that started the concept of NAIL-MS.
Oligonucleotide NAIL-MS
NAIL-MS can also be applied to oligonucleotide analysis by mass spectrometry. This is useful when the sequence information is to be retained.[11]
References
- ↑ Reichle, Valentin F.; Kaiser, Steffen; Heiss, Matthias; Hagelskamp, Felix; Borland, Kayla; Kellner, Stefanie (1 March 2019). "Surpassing limits of static RNA modification analysis with dynamic NAIL-MS". Methods. 156: 91–101. doi:10.1016/j.ymeth.2018.10.025. ISSN 1095-9130. PMID 30395967.
- ↑ Kellner, Stefanie; Neumann, Jennifer; Rosenkranz, David; Lebedeva, Svetlana; Ketting, René F.; Zischler, Hans; Schneider, Dirk; Helm, Mark (4 April 2014). "Profiling of RNA modifications by multiplexed stable isotope labelling". Chemical Communications. 50 (26): 3516–3518. doi:10.1039/c3cc49114e. ISSN 1364-548X. PMID 24567952.
- ↑ Dal Magro, Christina; Keller, Patrick; Kotter, Annika; Werner, Stephan; Duarte, Victor; Marchand, Virginie; Ignarski, Michael; Freiwald, Anja; Müller, Roman-Ulrich; Dieterich, Christoph; Motorin, Yuri (25 June 2018). "A Vastly Increased Chemical Variety of RNA Modifications Containing a Thioacetal Structure". Angewandte Chemie International Edition in English. 57 (26): 7893–7897. doi:10.1002/anie.201713188. ISSN 1521-3773. PMID 29624844.
- 1 2 Reichle, Valentin F.; Petrov, Dimitar P.; Weber, Verena; Jung, Kirsten; Kellner, Stefanie (6 December 2019). "NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli". Nature Communications. 10 (1): 5600. Bibcode:2019NatCo..10.5600R. doi:10.1038/s41467-019-13565-9. ISSN 2041-1723. PMC 6898146. PMID 31811240.
- ↑ Quinlivan, Eoin P.; Gregory, Jesse F. (15 February 2008). "DNA digestion to deoxyribonucleoside: a simplified one-step procedure". Analytical Biochemistry. 373 (2): 383–385. doi:10.1016/j.ab.2007.09.031. ISSN 0003-2697. PMC 2239294. PMID 18028864.
- ↑ Crain, P. F. (1990). Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods in Enzymology. Vol. 193. pp. 782–790. doi:10.1016/0076-6879(90)93450-y. ISSN 0076-6879. PMID 1706062.
- 1 2 Reichle, Valentin F.; Weber, Verena; Kellner, Stefanie (18 December 2018). "NAIL-MS in E. coli Determines the Source and Fate of Methylation in tRNA". ChemBioChem. 19 (24): 2575–2583. doi:10.1002/cbic.201800525. ISSN 1439-7633. PMC 6582434. PMID 30328661.
- ↑ Heiss, Matthias; Reichle, Valentin F.; Kellner, Stefanie (2 September 2017). "Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS". RNA Biology. 14 (9): 1260–1268. doi:10.1080/15476286.2017.1325063. ISSN 1555-8584. PMC 5699550. PMID 28488916.
- ↑ Heiss, Matthias; Hagelskamp, Felix; Marchand, Virginie; Motorin, Yuri; Kellner, Stefanie (15 January 2021). "Cell culture NAIL-MS allows insight into human tRNA and rRNA modification dynamics in vivo". Nature Communications. 12 (1): 389. doi:10.1038/s41467-020-20576-4. ISSN 2041-1723. PMC 7810713. PMID 33452242.
- ↑ Kellner, Stefanie; Ochel, Antonia; Thüring, Kathrin; Spenkuch, Felix; Neumann, Jennifer; Sharma, Sunny; Entian, Karl-Dieter; Schneider, Dirk; Helm, Mark (16 August 2014). "Absolute and relative quantification of RNA modifications via biosynthetic isotopomers". Nucleic Acids Research. 42 (18): e142. doi:10.1093/nar/gku733. ISSN 1362-4962. PMC 4191383. PMID 25129236.
- ↑ Hagelskamp, Felix; Borland, Kayla; Ramos, Jillian; Hendrick, Alan G.; Fu, Dragony; Kellner, Stefanie (21 February 2020). "Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro". Nucleic Acids Research. 48 (7): e41. doi:10.1093/nar/gkaa091. ISSN 1362-4962. PMC 7144906. PMID 32083657.