 | |
| Names | |
|---|---|
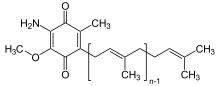
| IUPAC name
5-[(2E,6E,10E,14E,18E,22E,26E,30E,34E)-3,7,11,15,19,23,27,31,35,39-Decamethyltetraconta-2,6,10,14,18,22,26,30,34,38-decaenyl]-2-amino-3-methoxy-6-methylcyclohexa-2,5-diene-1,4-dione | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
PubChem CID |
|
| UNII | |
| |
| |
| Properties | |
| C58H89NO3 | |
| Molar mass | 848.354 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Rhodoquinone (RQ) is a modified ubiquinone-like molecule that is an important cofactor used in anaerobic energy metabolism by many organisms. Recently, it has gained attention as a potential anthelmintic drug target due to the fact that parasitic hosts do not synthesize or use this cofactor. Because this cofactor is used in low oxygen environments, many helminth-like organisms have adapted to survive host environments such as the areas within the gastrointestinal tracks.[1][2]
Biosynthesis
Currently the biosynthesis of rhodoquinone (RQ) is still being debated, but there are two main biosynthetic pathways that are being researched. The first pathway requires the organism to produce ubiquinone (UQ) before the amino group can be added onto the quinone ring. The second pathway allows RQ to be synthesized without any UQ being present by using tryptophan metabolites instead.[3]
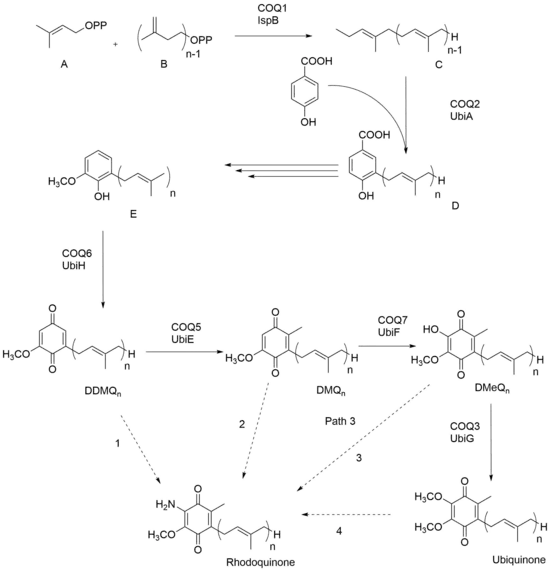
In the case of the prokaryotic organism R. rubrum, RQ has been shown to be synthesized by addition of an amino group to a pre-existing UQ; thus UQ needs to be present as a precursor before RQ can be made. Figure 1 shows the biosynthesis of UQ in yeast and E. coli where ‘n’ represents the number of isoprene units between various organisms. Dimethylallyl diphosphate A and isopentyl diphosphate B come together to form polyisoprenyl diphosphate C. With the addition of p-hydroxybenzoic acid, the product that arises is 3-polyprenyl-4-hydroxybenzoic acid D. The next three steps of synthesis varies between different organisms, but molecule E is made across all organisms and through oxidation, demethyldemethoxyubiquinone (DDMQ) is eventually formed. RQ has been theorized to be synthesized from DDMQn, DMQn, DMeQn, or UQn, as shown with the dashed arrows. Recent studies have shown that Path 4 - RQ biosynthesis via UQ, is the favored route.[4] It has been further shown that the gene rquA is required for the biosynthesis of RQ in R. rubrum, and that RquA catalyzes the conversion of UQ to RQ.[5][6] The RquA protein uses S-adenosyl-L-methionine as the amino donor to convert UQ to RQ in an unusual Mn(II)-catalyzed reaction.[7]
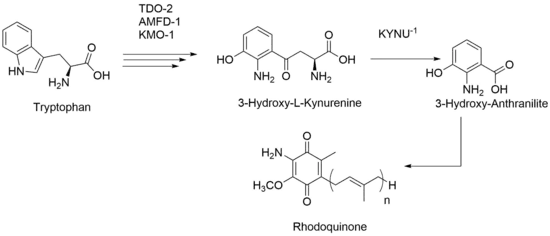
Research in C. elegans has shown an alternative path for production of RQ. Even after knocking out all UQ production, RQ is still present within those mutant strains. Based on this data, RQ production is not solely based on UQ-like molecules and instead can be made via tryptophan metabolites. Therefore, the amino group that is added in late stages of RQ biosynthesis in rquA-containing species is instead present throughout intermediate stages of RQ biosynthesis in C. elegans. With this proposed biosynthesis, the kynurenine pathway still needs to be upregulated, and activity from certain genes like kynu-1 which encodes for the KYNU-1 enzyme that catalyzes production of 3-hydroxy-L-kynurenine to 3-hydroxyanthranilic acid, needs to be upheld.[8][9] Recent work has revealed that alternative splicing of the coq-2 polyprenyltransferase gene controls the level of RQ in animals.[10] Animals that produce RQ (e.g. C. elegans and helminth parasites) contain both COQ-2 protein isoforms (COQ-2a and COQ-2e), and COQ-2e catalyzes prenylation of 3-hydroxyanthranilic acid (instead of p-hydroxybenzoic acid) which leads to RQ.
References
- ↑ "[Prevention and control of schistosomiasis and soil-transmitted helminthiasis:report of a WHO expert committee]". World Health Organization. 49 (3): 57. June 2012. ISBN 978-9241209120.
- ↑ Stairs CW, Eme L, Muñoz-Gómez SA, Cohen A, Dellaire G, Shepherd JN, et al. (April 2018). "Microbial eukaryotes have adapted to hypoxia by horizontal acquisitions of a gene involved in rhodoquinone biosynthesis". eLife. 7. doi:10.7554/eLife.34292. PMC 5953543. PMID 29697049.
- ↑ Salinas G, Langelaan DN, Shepherd JN (November 2020). "Rhodoquinone in bacteria and animals: Two distinct pathways for biosynthesis of this key electron transporter used in anaerobic bioenergetics". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1861 (11): 148278. doi:10.1016/j.bbabio.2020.148278. PMID 32735860.
- ↑ Brajcich BC, Iarocci AL, Johnstone LA, Morgan RK, Lonjers ZT, Hotchko MJ, et al. (January 2010). "Evidence that ubiquinone is a required intermediate for rhodoquinone biosynthesis in Rhodospirillum rubrum". Journal of Bacteriology. 192 (2): 436–445. doi:10.1128/JB.01040-09. PMC 2805321. PMID 19933361.
- ↑ Lonjers ZT, Dickson EL, Chu TP, Kreutz JE, Neacsu FA, Anders KR, Shepherd JN (March 2012). "Identification of a new gene required for the biosynthesis of rhodoquinone in Rhodospirillum rubrum". Journal of Bacteriology. 194 (5): 965–971. doi:10.1128/JB.06319-11. PMC 3294814. PMID 22194448.
- ↑ Bernert AC, Jacobs EJ, Reinl SR, Choi CC, Roberts Buceta PM, Culver JC, et al. (September 2019). "Recombinant RquA catalyzes the in vivo conversion of ubiquinone to rhodoquinone in Escherichia coli and Saccharomyces cerevisiae". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1864 (9): 1226–1234. doi:10.1016/j.bbalip.2019.05.007. PMC 6874216. PMID 31121262.
- ↑ Neupane T, Chambers LR, Godfrey AJ, Monlux MM, Jacobs EJ, Whitworth S, et al. (August 2022). "Microbial rhodoquinone biosynthesis proceeds via an atypical RquA-catalyzed amino transfer from S-adenosyl-L-methionine to ubiquinone". Communications Chemistry. 5 (1): 89. doi:10.1038/s42004-022-00711-6. PMC 9814641. PMID 36697674.
- ↑ Roberts Buceta PM, Romanelli-Cedrez L, Babcock SJ, Xun H, VonPaige ML, Higley TW, et al. (July 2019). "The kynurenine pathway is essential for rhodoquinone biosynthesis in Caenorhabditis elegans". The Journal of Biological Chemistry. 294 (28): 11047–11053. doi:10.1074/jbc.AC119.009475. PMC 6635453. PMID 31177094.
- ↑ Del Borrello S, Lautens M, Dolan K, Tan JH, Davie T, Schertzberg MR, et al. (June 2019). "Rhodoquinone biosynthesis in C. elegans requires precursors generated by the kynurenine pathway". eLife. 8. doi:10.7554/eLife.48165. PMC 6656428. PMID 31232688.
- ↑ Tan JH, Lautens M, Romanelli-Cedrez L, Wang J, Schertzberg MR, Reinl SR, et al. (August 2020). "Alternative splicing of coq-2 controls the levels of rhodoquinone in animals". eLife. 9: e56376. doi:10.7554/eLife.56376. PMC 7434440. PMID 32744503.