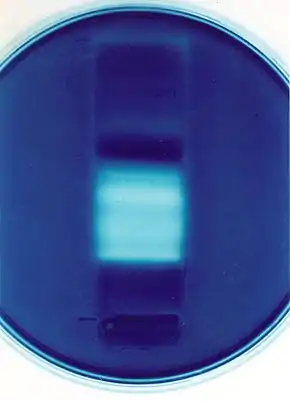
Zymography is an electrophoretic technique for the detection of hydrolytic enzymes, based on the substrate repertoire of the enzyme. Three types of zymography are used; in gel zymography, in situ zymography and in vivo zymography.[2] For instance, gelatin embedded in a polyacrylamide gel will be digested by active gelatinases run through the gel. After Coomassie staining, areas of degradation are visible as clear bands against a darkly stained background.[3]
Modern usage of the term zymography has been adapted to define the study and cataloging of fermented products, such as beer or wine, often by specific brewers or winemakers or within an identified category of fermentation such as with a particular strain of yeast or species of bacteria. Zymography also refers to a collection of related, fermented products, considered as a body of work. For example, all of the beers produced by a particular brewery could collectively be referred to as its zymography.
See also Zymology or the applied science of zymography. Zymology relates to the biochemical processes of fermentation, especially the selection of fermenting yeast and bacteria in brewing, winemaking, and other fermented foods. For example, beer-making involves the application of top (ale) or bottom fermenting yeast (lager), to produce the desired variety of beer. The synthesis of the yeast can impact the flavor profile of the beer, i.e. diacetyl (taste or aroma of buttery, butterscotch).
Gel zymography
Samples are prepared in a standard, non-reducing loading buffer for SDS-PAGE. No reducing agent or boiling are necessary since these would interfere with refolding of the enzyme. A suitable substrate (e.g. gelatin or casein for protease detection) is embedded in the resolving gel during preparation of the acrylamide gel. Following electrophoresis, the SDS is removed from the gel (or zymogram) by incubation in unbuffered Triton X-100, followed by incubation in an appropriate digestion buffer, for an optimized length of time at 37 °C. The zymogram is subsequently stained (commonly with Amido Black or Coomassie brilliant blue), and areas of digestion appear as clear bands against a darkly stained background where the substrate has been degraded by the enzyme.
Variations on the standard protocol
The standard protocol may require modifications depending on the sample enzyme; for instance, D. melanogaster digestive glycosidases generally survive reducing conditions (i.e. the presence of 2-mercaptoethanol or DTT), and to an extent, heating. Indeed, the separations following heating to 50 °C tend to exhibit a substantial increase in band resolution, without appreciable loss of activity.[4][5]
A common protocol used in the past for zymography of α-amylase activity was the so-called starch film protocol of W.W. Doane. Here a native PAGE gel was run to separate the proteins in a homogenate. Subsequently, a thin gel with starch dissolved (or more properly, suspended) in it was overlaid for a period of time on top of the original gel.[6] The starch was then stained with Lugol's iodine.
Gel zymography is often used for the detection and analysis of enzymes produced by microorganisms.[7] This has led to variations on the standard protocol e.g. mixed-substrate zymography.[2]
Reverse zymography copolymerizes both the substrate and the enzyme with the acrylamide, and is useful for the demonstration of enzyme inhibitor activity. Following staining, areas of inhibition are visualized as dark bands against a clear (or lightly stained) background.
In imprint technique, the enzyme is separated by native gel electrophoresis and the gel is laid on top of a substrate treated agarose.[1]
Zymography can also be applied to other types of enzymes, including xylanases, lipases and chitinases.
See also
References
- 1 2 Hempelmann, E.; Putfarken, B.; Rangachari, K.; Wilson, R.J.M. (1986). "Immunoprecipitation of malarial acid endopeptidase". Parasitology. 92 (2): 305–312. doi:10.1017/S0031182000064076. PMID 3520446. S2CID 42414630.
- 1 2 Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G (2013). "Zymography methods for visualizing hydrolytic enzymes". Nat Methods. 10 (3): 211–220. doi:10.1038/nmeth.2371. PMID 23443633. S2CID 5314901.
- ↑ "Gelatin zymography protocol | Abcam". www.abcam.com. Retrieved 2017-05-12.
- ↑ Martínez TF, Alarcón FJ, Díaz-López M, Moyano FJ (2000). "Improved detection of amylase activity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with copolymerized starch". Electrophoresis. 21 (14): 2940–2943. doi:10.1002/1522-2683(20000801)21:14<2940::AID-ELPS2940>3.0.CO;2-S. PMID 11001307. S2CID 30732170.
- ↑ Snoek-van Beurden PA, Von den Hoff JW (2005). "Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors". BioTechniques. 38 (1): 73–83. doi:10.2144/05381RV01. hdl:2066/47379. PMID 15679089.
- ↑ Doane WW (1969). "Amylase variants in Drosophila melanogaster: linkage studies and characterization of enzyme extracts". J Exp Zool. 171 (3): 31–41. doi:10.1002/jez.1401710308. PMID 5348624.
- ↑ Lantz MS, Ciborowski P (1994). "Zymographic techniques for detection and characterization of microbial proteases". Methods Enzymol. Methods in Enzymology. 235: 563–594. doi:10.1016/0076-6879(94)35171-6. ISBN 9780121821364. PMID 8057927.