| HLA-A33 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
 HLA-A33 | ||||||||||||||||
| About | ||||||||||||||||
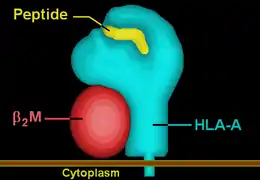
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*33--, β2-microglobulin | |||||||||||||||
| Subtypes | ||||||||||||||||
| ||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
HLA-A33 (A33) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α33 subset of HLA-A α-chains. For A33, the alpha "A" chain are encoded by the HLA-A*33 allele group and the β-chain are encoded by B2M locus.[1] A33 and A*33 are almost synonymous in meaning. A33 is a split antigen of the broad antigen serotype A19. A33 is a sister serotype of A29, A30, A31, A32, and A74.
A33 is more common in Subsaharan Africa.
Serotype
| A*33 | A33 | A19 | Sample |
| allele | % | % | size (N) |
| *3301 | 87 | 3 | 687 |
| *3303 | 95 | 0 | 807 |
A33 has a poor serotyping rate.
A33 frequencies
| freq | ||
| ref. | Population | (%) |
| [3] | Pakistan Karachi Parsi | 12.2 |
| [4] | Portugal North | 7.6 |
| [5] | Tunisia | 6.1 |
| [6] | Morocco Nador Metalsa | 5.5 |
| [4] | Mongolia Buriat | 4.6 |
| [7] | Guinea Bissau | 4.6 |
| [8] | Jordan Amman | 3.5 |
| [7] | Cape Verde NW Islands | 3.2 |
| [4] | Portugal Centre | 3.0 |
| [9] | Iran Baloch | 2.8 |
| [10] | Pakistan Baloch | 2.4 |
| [4] | France South East | 2.3 |
| [4] | Sudanese | 1.8 |
| [4] | Japan Okinawa Ryukyuan | 1.8 |
| [11] | Mali Bandiagara | 1.8 |
| [12] | Bulgaria | 1.8 |
| [10] | Pakistan Kalash | 1.7 |
| [11] | Uganda Kampala | 1.5 |
| [13] | Georgia Svaneti | 1.3 |
| [11] | Kenya Luo | 1.3 |
| [14] | Oman | 1.3 |
| [11] | Zambia Lusaka | 1.2 |
| [15] | China Guangdong Meizhou | 1.0 |
| [4] | Romanian | 1.0 |
| [4] | Croatia | 1.0 |
| [16] | China Qinghai Hui | 0.9 |
| [4] | Czech Republic[17] | 0.9 |
| [4] | Kenya | 0.7 |
| [18] | Ireland Northern | 0.7 |
| [14] | Singapore Chinese | 0.6 |
| [19] | Cameroon Bamileke | 0.6 |
| [20] | India North Delhi | 0.5 |
| [4] | Georgia Tbilisi | 0.5 |
| [4] | Belgium | 0.5 |
| [19] | Cameroon Beti | 0.3 |
| [11] | Kenya Nandi | 0.2 |
A33 shows two different distributions that can be discriminated by subtyping capability of SSP-PCR.
A*3301 distribution
The first distribution appears to have a Western distribution that introgresses into Europe as a result of the Post-neolithic periods. It is commonly found in linkage disequilibration within the A*3301-Cw*0802-B*1402 haplotype which can be extended to DRB1 and DQB1 in certain instances (See Below). The source of its general expansion appears to be the middle east or the levant, as it is found in the Palestinian population. B14 splits into B64 (B*1401) and B65 (B*1402) but the only Arabian people which show both antigens are the United Arab Emirates.
A*3303 distribution
| freq | ||
| ref. | Population | (%) |
| [19] | Cameroon Baka | 25.0 |
| [19] | Cameroon Sawa | 23.1 |
| [4] | India West Bhils | 18.0 |
| [10] | Pakistan Burusho | 17.9 |
| [21] | South Korea (3) | 16.3 |
| [4] | India West Parsis | 14.0 |
| [4] | Singapore Thai | 13.3 |
| [4] | India Mumbai Marathas | 13.0 |
| [22] | Japan | 12.8 |
| [10] | Pakistan Baloch | 12.7 |
| [4] | China South Han | 11.5 |
| [4] | Singapore Riau Malay | 10.9 |
| [14] | Singapore Chinese | 10.1 |
| [4] | Hong Kong Chinese | 10.0 |
| [23] | Chaoshan | 9.8 |
| [11] | Mali Bandiagara | 9.4 |
| [16] | China Inner Mongolia | 9.3 |
| [4] | Singapore Chinese Han | 9.3 |
| [4] | Singapore Javan | 9.0 |
| [24] | India North Hindus | 8.7 |
| [7] | Guinea Bissau | 8.5 |
| [4] | Taiwan Minnan | 8.3 |
| [4] | Taiwan Hakka | 8.2 |
| [25] | Senegal Mandenka | 8.1 |
| [10] | Pakistan Brahui | 8.0 |
| [10] | Pakistan Sindhi | 7.6 |
| [4] | Russia Tuvan | 7.1 |
| [10] | Pakistan Pathan | 7.1 |
| [4] | South Africa Natal Tamil | 7.0 |
| [3] | Pakistan Karachi Parsi | 6.7 |
| [26] | India Tamil Nadu Nadar | 6.6 |
| [4] | Israel Jews | 6.4 |
| [4] | India Andhra Pradesh Golla | 6.3 |
| [27] | China North Han | 6.2 |
| [28] | China Beijing Tianjian | 6.2 |
| [20] | India New Delhi | 6.1 |
| [10] | Pakistan Kalash | 5.8 |
| [9] | Iran Baloch | 5.6 |
| [16] | China Qinghai Hui | 5.5 |
| [4] | China Guangzhou | 5.4 |
| [19] | Cameroon Bamileke | 4.5 |
| [29] | China Yunnan Nu | 4.4 |
| [4] | China Guangxi Maonan | 4.2 |
| [15] | China Guangdong Meizhou | 4.0 |
| [7] | Cape Verde SW Islands | 4.0 |
| [4] | Taiwan Pazeh | 3.6 |
| [4] | Uganda Kampala | 3.1 |
| [4] | China Beijing | 3.0 |
| [4] | India Khandesh Pawra | 3.0 |
| [19] | Cameroon Yaounde | 2.8 |
| [4] | Sudanese | 2.5 |
| [4] | Zimbabwe Harare Shona | 2.4 |
| [7] | Cape Verde NW Islands | 2.4 |
| [19] | Cameroon Beti | 2.3 |
| [14] | Oman | 2.1 |
| [30] | China Tibet | 1.6 |
| [4] | Croatia | 1.3 |
| [4] | Romanian | 1.2 |
| [29] | China Yunnan Lisu | 1.1 |
| [4] | Arab Druse | 1.0 |
| [4] | Georgia Tbilisi | 1.0 |
| [4] | Belgium | 1.0 |
| [11] | Kenya Luo | 0.9 |
| [31] | Lakota Sioux | 0.5 |
| [4] | Australian Indig. Cape York | 0.5 |
| [5] | Tunisia | 0.5 |
| [14] | South African Natal Zulu | 0.5 |
| [4] | Kenya | 0.3 |
| [18] | Ireland Northern | 0.3 |
| [11] | Kenya Nandi | 0.2 |
Certain alleles confound population histories. At the top of that list is A*3303. This allele appears to jump, figuratively, out of West Africa into South Asia. The point of origin is Africa, most likely central or western Africa given the low levels in East Africa (although much of East Africa is undersampled). In certain tested populations of the Middle East the leve of A*3303 is either very low, or non-existent. Within East Africa Sudan appears to be the highest at around 2%. The frequency of A*3303 begins to rise in eastern Arabia (Oman, UAE) and then markedly rise in the Brahui and Balochi of Pakistan. One haplotype stands out, the A33-B58-DR3-DQ2 haplotype which is found in West Africa, in Sudan, and Pakistan, scattered along West Indias coast, the Turkic republics and appears to have recently introgressed into Korea (post-Yayoi period of Japan) and China. So recent arrival into Asia that the level of HLA DR3-DQ2 in Korea of 2.9%. Korea is the major recent source of Japanese genes, by the Yayoi period that lasted from 3000 to 1600 years ago approximately 3/4ths of Japanese genetic makeup is attributed to this migration. And yet there is trace DR3-DQ2 in Japanese, none in the Ainu nor many other indigenous Siberian groups.
A33 Haplotypes
A33-Cw8-B14-DR1-DQ5
| freq | ||
| ref. | Population | (%) |
| [3] | Parsis (Pakistan) | 4.4 |
| [32] | Sardinian | 3.0 |
| India West Coast Parsis | 3.3[33] | |
| [32] | Portuguese | 2.7 |
| [32] | Armenian | 2.5 |
| [32] | Indian | 2.0 |
| [32] | Polish | 2.0 |
| Ashkenazi Jews | 1.8 | |
| [32] | French | 1.8 |
| [32] | Spanish | 1.7 |
| [32] | Albanian | 1.5 |
| [32] | German | 1.5 |
| [32] | Tuscan | 1.3 |
| [32] | Greek | 1.1 |
| [32] | Marathans | 1.1 |
| [32] | Italian | 1.0 |
When dealing with haplotypes, if one assumes that linkage disequilibrium is random, then one can estimate the time of equilibration based on the size of the haplotype, the A-B-DR haplotype is over 2 million nucleotides in length. Given this length it is unlikely it spread during the Neolithic period. A more like guess as to when it spread was the early historic period, with the spread of the Phoenician and Mycenaean culture throughout the mediterranean. Its presence in India, particularly northern India, indicates possible spread of this haplotype within the Black Sea region prior to the migration of Indo-Aryan culture across the Indus River. The specific nomenclature for this type is:
A*3301 : C*0802 : B*1402 : DRB1*0102 : DQA1*0102 : DQB1*0501
A33-B44
| freq | ||
| ref. | Population | (%) |
| Iyers | 9.6 | |
| Korea | 8.0 | |
| Japan | 6.1 | |
| Thais | 4.7 | |
| [3] | Parsis (Pakistan) | 4.4 |
| Bharghavas | 4.0 | |
| Java | 3.7 | |
| Tribals (India) | 3.0 | |
| Chinese (Thailand) | 2.8 | |
| Vietnamese | 2.7 |
This haplotype appears to precede A33-B58 in Asia, bringing with it the DR7-DQ2 haplotype. There are two versions of the haplotype, possibly of different origins. It's a good reason why serotyping alone should not be relied upon. The first haplotype is A33-Cw14-B44-DR13-DQ6.4[34]
A*3303 : C*1403 : B*4403 : DRB1*1302 : DQA1*0102 : DQB1*0604 : DPB1*0401
This haplotype is found in Japan and Korea, and it is the most common 5 locus HLA type in Korea, high at 4.2%, 25 times higher than in China. In Japan it is 4.8% and can be extended to DPB1 at 3.6%. While clearly not showing the level of disequilibrium of the Super B8 haplotype, the level of disequilibrium is high, indicating an expansive migration into these regions at some time in the recent past, most likely in the period preceding the Yayoi period of Japan.
A*3303 : C*0701 : B*4403 : DRB1*0701 : DQA1*0201? : DQB1*0202
The second haplotype, like A33-B58 is found in Korea but not in Japan.[21] This haplotype carries the other common DQ2 haplotype, DQ2.2. The Cw*0701 is found in the A*33-B58 haplotype and is like the result of a recombination between A33-Cw7 and a different B44-DR7 haplotype. These haplotypes indicate that interpreting population relationships by allele or even by low resolution haplotype information is error-prone and suggests the need for high resolution multigene haplotype studies.
A33-Cw3-B58-DR3-DQ2
| freq | ||
| ref. | Population | (%) |
| [32] | Chinese (Thailand) | 12.6 |
| [10] | Baloch (Pakistan) | 11.1 |
| [23] | Chaoshan (China) | 8.1 |
| [32] | Chinese (Singapore) | 5.5 |
| [10] | Burusho (Pakistan) | 4.6 |
| [32] | Hui | 4.0 |
| [32] | Mongolian | 3.7 |
| [10] | Kalash (Pakistan) | 3.6 |
| [32] | Korea | 3.5 |
| [32] | Yaku | 3.2 |
| [10] | Panthan (Pakistan) | 3.0 |
| [32] | Tribals (India) | 3.0 |
| Baloch (SE Iran) | 2.9 | |
| [10] | Brahui (Pakistan) | 2.9 |
| [32] | Southern Han | 2.8 |
| [32] | Thais | 2.5 |
| [32] | Vietnamese | 2.3 |
| [32] | Inner Mongolian | 2.2 |
| [32] | Miao | 2.1 |
| [32] | West African | 2.1 |
| [32] | South African | 1.9 |
| [35] | Oman | 1.6 |
| [10] | Sindhi (Pakistan) | 1.5 |
| [32] | Manchu | 1.2 |
| [36] | Sudan | 1.2 |
| Cameroon Yaounde[37] | 1.1 |
Within eastern Asia A*3303 is in linkage disequilibrium with on haplotype in particular, the specific genetic makeup is:
A*3303 : C*0302 : B*5801 : DRB1*0301 : DQA1*0501 : DQB1*0201
It is interesting that the Cw allele in the Pakistani population is the same as the allele in the east Asian population C*0302. 8.3 of 11.1% of the A33-B58 in the Baloch Pakistani can is linked to DR3 and presumably DQ2.5 (There are few exceptions outside of Africa). This extends a haplotype the forms a semicircle around the Indian subcontinent indicating a substantive and relatively recent genetic relationship. The Parsis of Pakistan lack A33-B58, as with groups to the far west of Pakistan. The A33-B58-DR3-DQ2 haplotype appears to have originated in whole from West Africa, with current possibilities for Sudan or Northern Ethiopia as points of exit from Africa and a migration by the Indian Ocean to the western side of the Indus River.
A33-Cw7-B58-DR13-DQ6
Within eastern Asia A*3303 is in linkage disequilibrium with on haplotype in particular, the specific genetic makeup is:
A*3303 : C*0701 : B*5801 : DRB1*1302 : DQA1*0102 : DQB1*0609
This haplotype is composed of genes most frequent in parts of western Africa. This includes the A*3303, B*5801, DRB1*1302, and DQB1*0609. The DRB1*0609 haplotype in nodal in east/central Africa in the Ugandan, Rwanda, Congo, Cameroon whereas the allele is at low frequencies in Western Europe, and its distribution is also consistent with a migration from east Africa direct to the Lower Indus River.
References
- ↑ Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- ↑ Allele Query Form IMGT/HLA - European Bioinformatics Institute
- 1 2 3 4 Mohyuddin A, Mehdi SQ (2005). "HLA analysis of the Parsi (Zoroastrian) population in Pakistan". Tissue Antigens. 66 (6): 691–5. doi:10.1111/j.1399-0039.2005.00507.x. PMID 16305686.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–7. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
{{cite journal}}: External link in|title= - 1 2 Ayed K, Ayed-Jendoubi S, Sfar I, Labonne MP, Gebuhrer L (2004). "HLA class-I and HLA class-II phenotypic, gene and haplotypic frequencies in Tunisians by using molecular typing data". Tissue Antigens. 64 (4): 520–32. doi:10.1111/j.1399-0039.2004.00313.x. PMID 15361135.
- ↑ Piancatelli D, Canossi A, Aureli A, et al. (2004). "Human leukocyte antigen-A, -B, and -Cw polymorphism in a Berber population from North Morocco using sequence-based typing". Tissue Antigens. 63 (2): 158–72. doi:10.1111/j.1399-0039.2004.00161.x. PMID 14705987.
- 1 2 3 4 5 Spínola H, Bruges-Armas J, Middleton D, Brehm A (2005). "HLA polymorphisms in Cabo Verde and Guiné-Bissau inferred from sequence-based typing". Hum. Immunol. 66 (10): 1082–92. doi:10.1016/j.humimm.2005.09.001. PMID 16386651.
- ↑ Sánchez-Velasco P, Karadsheh NS, García-Martín A, Ruíz de Alegría C, Leyva-Cobián F (2001). "Molecular analysis of HLA allelic frequencies and haplotypes in Jordanians and comparison with other related populations". Hum. Immunol. 62 (9): 901–9. doi:10.1016/S0198-8859(01)00289-0. PMID 11543892.
- 1 2 Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H (2004). "Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of Pakistan". Tissue Antigens. 64 (5): 581–7. doi:10.1111/j.1399-0039.2004.00302.x. PMID 15496201.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Mohyuddin A, Ayub Q, Qamar R, Khaliq S, Mansoor A, Mehdi SQ (1999). "HLA polymorphisms in ethnic groups from Pakistan". Transplant. Proc. 31 (8): 3350–1. doi:10.1016/S0041-1345(99)00821-0. PMID 10616502.
- 1 2 3 4 5 6 7 8 Cao K, Moormann AM, Lyke KE, et al. (2004). "Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci". Tissue Antigens. 63 (4): 293–325. doi:10.1111/j.0001-2815.2004.00192.x. PMID 15009803.
- ↑ Ivanova M, Rozemuller E, Tyufekchiev N, Michailova A, Tilanus M, Naumova E (2002). "HLA polymorphism in Bulgarians defined by high-resolution typing methods in comparison with other populations". Tissue Antigens. 60 (6): 496–504. doi:10.1034/j.1399-0039.2002.600605.x. PMID 12542743.
- ↑ Sánchez-Velasco P, Leyva-Cobián F (2001). "The HLA class I and class II allele frequencies studied at the DNA level in the Svanetian population (Upper Caucasus) and their relationships to Western European populations". Tissue Antigens. 58 (4): 223–33. doi:10.1034/j.1399-0039.2001.580402.x. PMID 11782273.
- 1 2 3 4 5 Middleton D, Williams F, Meenagh A, et al. (2000). "Analysis of the distribution of HLA-A alleles in populations from five continents". Hum. Immunol. 61 (10): 1048–52. doi:10.1016/S0198-8859(00)00178-6. PMID 11082518.
- 1 2 Chen S, Li W, Hu Q, Liu Z, Xu Y, Xu A (2007). "Polymorphism of HLA class I genes in Meizhou Han population of Guangdong, China". International Journal of Immunogenetics. 34 (2): 131–6. doi:10.1111/j.1744-313X.2007.00669.x. PMID 17373939. S2CID 25192660.
- 1 2 3 Hong W, Chen S, Shao H, Fu Y, Hu Z, Xu A (2007). "HLA class I polymorphism in Mongolian and Hui ethnic groups from Northern China". Hum. Immunol. 68 (5): 439–48. doi:10.1016/j.humimm.2007.01.020. PMID 17462512.
- ↑ Bendukidze N, Ivasková E, Zahlavová L, et al. (2003). "Identification of HLA alleles with low or no cell surface expression in the Czech population". Folia Biol. (Praha). 49 (6): 227–9. PMID 14748437.
- 1 2 Williams F, Meenagh A, Maxwell AP, Middleton D (1999). "Allele resolution of HLA-A using oligonucleotide probes in a two-stage typing strategy". Tissue Antigens. 54 (1): 59–68. doi:10.1034/j.1399-0039.1999.540107.x. PMID 10458324.
- 1 2 3 4 5 6 7 Torimiro JN, Carr JK, Wolfe ND, et al. (2006). "HLA class I diversity among rural rainforest inhabitants in Cameroon: identification of A*2612-B*4407 haplotype". Tissue Antigens. 67 (1): 30–7. doi:10.1111/j.1399-0039.2005.00527.x. PMID 16451198.
- 1 2 Rani R, Marcos C, Lazaro AM, Zhang Y, Stastny P (2007). "Molecular diversity of HLA-A, -B and -C alleles in a North Indian population as determined by PCR-SSOP". International Journal of Immunogenetics. 34 (3): 201–8. doi:10.1111/j.1744-313X.2007.00677.x. PMID 17504510. S2CID 24547295.
- 1 2 Lee KW, Oh DH, Lee C, Yang SY (2005). "Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population". Tissue Antigens. 65 (5): 437–47. doi:10.1111/j.1399-0039.2005.00386.x. PMID 15853898.
- ↑ Tokunaga K, Ishikawa Y, Ogawa A, et al. (1997). "Sequence-based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes". Immunogenetics. 46 (3): 199–205. doi:10.1007/s002510050262. PMID 9211745. S2CID 22302364.
- 1 2 Hu SP, Luan JA, Li B, et al. (2007). "Genetic link between Chaoshan and other Chinese Han populations: Evidence from HLA-A and HLA-B allele frequency distribution". Am. J. Phys. Anthropol. 132 (1): 140–50. doi:10.1002/ajpa.20460. PMID 16883565.
- ↑ Rajalingam R, Krausa P, Shilling HG, et al. (2002). "Distinctive KIR and HLA diversity in a panel of north Indian Hindus". Immunogenetics. 53 (12): 1009–19. doi:10.1007/s00251-001-0425-5. PMID 11904677. S2CID 20035835.
- ↑ Sanchez-Mazas A, Steiner QG, Grundschober C, Tiercy JM (2000). "The molecular determination of HLA-Cw alleles in the Mandenka (West Africa) reveals a close genetic relationship between Africans and Europeans". Tissue Antigens. 56 (4): 303–12. doi:10.1034/j.1399-0039.2000.560402.x. PMID 11098930.
- ↑ Shankarkumar U, Sridharan B, Pitchappan RM (2003). "HLA diversity among Nadars, a primitive Dravidian caste of South India". Tissue Antigens. 62 (6): 542–7. doi:10.1046/j.1399-0039.2003.00118.x. PMID 14617038.
- ↑ Hong W, Fu Y, Chen S, Wang F, Ren X, Xu A (2005). "Distributions of HLA class I alleles and haplotypes in Northern Han Chinese". Tissue Antigens. 66 (4): 297–304. doi:10.1111/j.1399-0039.2005.00474.x. PMID 16185325.
- ↑ Yang G, Deng YJ, Hu SN, et al. (2006). "HLA-A, -B, and -DRB1 polymorphism defined by sequence-based typing of the Han population in Northern China". Tissue Antigens. 67 (2): 146–52. doi:10.1111/j.1399-0039.2006.00529.x. PMID 16441486.
- 1 2 Chen S, Hu Q, Xie Y, et al. (2007). "Origin of Tibeto-Burman speakers: evidence from HLA allele distribution in Lisu and Nu inhabiting Yunnan of China". Hum. Immunol. 68 (6): 550–9. doi:10.1016/j.humimm.2007.02.006. PMID 17509456.
- ↑ Chen S, Hong W, Shao H, et al. (2006). "Allelic distribution of HLA class I genes in the Tibetan ethnic population of China". International Journal of Immunogenetics. 33 (6): 439–45. doi:10.1111/j.1744-313X.2006.00637.x. PMID 17117954. S2CID 34168713.
- ↑ Leffell MS, Fallin MD, Hildebrand WH, Cavett JW, Iglehart BA, Zachary AA (2004). "HLA alleles and haplotypes among the Lakota Sioux: report of the ASHI minority workshops, part III". Hum. Immunol. 65 (1): 78–89. doi:10.1016/j.humimm.2003.10.001. PMID 14700599.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 Sasazuki, Takehiko; Tsuji, Kimiyoshi; Aizawa, Miki (1992). HLA 1991: proceedings of the eleventh International Histocompatibility Workshop and Conference, held in Yokohama, Japan, 6-13 November, 1991. Oxford [Oxfordshire]: Oxford University Press. ISBN 0-19-262390-7.
- ↑ bears the C*0502 instead of the C*0802 otherwise seen with this haplotype
- ↑ Saito S, Ota S, Yamada E, Inoko H, Ota M (2000). "Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population". Tissue Antigens. 56 (6): 522–9. doi:10.1034/j.1399-0039.2000.560606.x. PMID 11169242.
- ↑ Williams F, Meenagh A, Darke C, et al. (2001). "Analysis of the distribution of HLA-B alleles in populations from five continents". Hum. Immunol. 62 (6): 645–50. doi:10.1016/S0198-8859(01)00247-6. PMID 11390040.
- ↑ Ward FE, Jensen JB, Abdul Hadi NH, Stewart A, Vande Waa JV, Bayoumi RA (1989). "HLA genotypes and variant alleles in Sudanese families of Arab-Negroid tribal origin". Hum. Immunol. 24 (4): 239–51. doi:10.1016/0198-8859(89)90018-9. PMID 2708086.
- ↑ bears the A*3303:B*5802