Retroviral matrix proteins are components of envelope-associated capsids of retroviruses. These proteins line the inner surface of viral envelopes and are associated with viral membranes.[1]
Matrix proteins are produced as N-terminal domains of Gag precursor polyproteins. The Gag polyprotein directs the assembly and release of virus particles from infected cells. The Gag polyprotein has three domains required for activity: an N-terminal membrane-binding (M) domain (which corresponds to the matrix protein) that directs Gag to the plasma membrane, an interaction (I) domain involved in Gag aggregation, and a late assembly (L) domain that mediates the budding process .[2] During viral maturation, the Gag polyprotein is cleaved by the retroviral protease into several corresponding structural proteins, yielding the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, and some smaller peptides. Gag-derived proteins govern the entire assembly and release of the virus particles, with matrix proteins playing key roles in Gag stability, capsid assembly, transport and budding.
Families of retroviral matrix proteins
| Gag_MA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
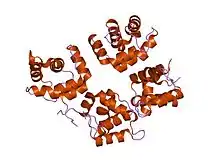 structure of moloney murine leukaemia virus matrix protein | |||||||||
| Identifiers | |||||||||
| Symbol | Gag_MA | ||||||||
| Pfam | PF01140 | ||||||||
| Pfam clan | CL0074 | ||||||||
| InterPro | IPR000840 | ||||||||
| SCOP2 | 1mn8 / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 42 | ||||||||
| OPM protein | 1mn8 | ||||||||
| |||||||||
Although matrix proteins from different viruses appear to perform similar functions and can have similar structural folds, their primary sequences can be very different. Typical matrix proteins of retroviruses form an alpha helical bundle structure .[3]
One family of these proteins represents matrix proteins from gammaretroviruses, such as Moloney murine leukemia virus (MoMLV), feline leukemia virus (FLV), and feline sarcoma virus (FESV).[4][5] This family also includes matrix proteins from several eukaryotic endogenous retroviruses, which arise when one or more copies of the retroviral genome becomes integrated into the host genome.[6]
| Retro_M | |||||||||
|---|---|---|---|---|---|---|---|---|---|
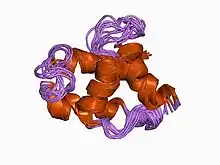 m-domain from gag polyprotein of rous sarcoma virus, nmr, 20 structures | |||||||||
| Identifiers | |||||||||
| Symbol | Retro_M | ||||||||
| Pfam | PF02813 | ||||||||
| Pfam clan | CL0074 | ||||||||
| InterPro | IPR004028 | ||||||||
| SCOP2 | 1a6s / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 42 | ||||||||
| OPM protein | 1a6s | ||||||||
| |||||||||
Another family represents the M domain of the Gag polyprotein found in avian retroviruses. It includes Gag polyproteins from several avian endogenous retroviruses.[7]
| Gag_p10 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 solution structure of the m-pmv wild type matrix protein (p10) | |||||||||
| Identifiers | |||||||||
| Symbol | Gag_p10 | ||||||||
| Pfam | PF02337 | ||||||||
| Pfam clan | CL0074 | ||||||||
| InterPro | IPR003322 | ||||||||
| OPM superfamily | 44 | ||||||||
| OPM protein | 2lpy | ||||||||
| |||||||||
Matrix proteins are also components of beta-retroviruses such as Mason-Pfizer monkey virus (MPMV) and mouse mammary tumor virus (MMTV) .[8][9] This entry also identifies matrix proteins from several eukaryotic endogenous retroviruses.[6]
| Gag_p15 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
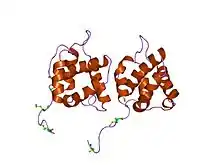 crystal structure of equine infectious anaemia virus matrix antigen (eiav ma) | |||||||||
| Identifiers | |||||||||
| Symbol | Gag_p15 | ||||||||
| Pfam | PF08723 | ||||||||
| InterPro | IPR014834 | ||||||||
| SCOP2 | 1hek / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 44 | ||||||||
| OPM protein | 1hek | ||||||||
| |||||||||
References
- ↑ Conte MR, Matthews S (July 1998). "Retroviral matrix proteins: a structural perspective". Virology. 246 (2): 191–8. doi:10.1006/viro.1998.9206. PMID 9657938.
- ↑ Parent LJ, Cairns TM, Albert JA, Wilson CB, Wills JW, Craven RC (January 2000). "RNA dimerization defect in a Rous sarcoma virus matrix mutant". J. Virol. 74 (1): 164–72. doi:10.1128/jvi.74.1.164-172.2000. PMC 111525. PMID 10590103.
- ↑ McDonnell JM, Fushman D, Cahill SM, Zhou W, Wolven A, Wilson CB, Nelle TD, Resh MD, Wills J, Cowburn D (June 1998). "Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus". J. Mol. Biol. 279 (4): 921–8. doi:10.1006/jmbi.1998.1788. PMID 9642071.
- ↑ Riffel N, Harlos K, Iourin O, Rao Z, Kingsman A, Stuart D, Fry E (December 2002). "Atomic resolution structure of Moloney murine leukemia virus matrix protein and its relationship to other retroviral matrix proteins". Structure. 10 (12): 1627–36. doi:10.1016/S0969-2126(02)00896-1. PMID 12467570.
- ↑ Baker SJ, Cosenza SC, Reddy EP (September 1998). "The role of v-Fgr myristoylation and the Gag domain in membrane binding and cellular transformation". Virology. 249 (1): 1–11. doi:10.1006/viro.1998.9323. PMID 9740771.
- 1 2 Gifford R, Tristem M (May 2003). "The evolution, distribution and diversity of endogenous retroviruses". Virus Genes. 26 (3): 291–315. doi:10.1023/A:1024455415443. PMID 12876457. S2CID 34639116.
- ↑ Borisenko L (2003). "Avian endogenous retroviruses". Folia Biol. (Praha). 49 (5): 177–82. PMID 14680291.
- ↑ Stansell E, Tytler E, Walter MR, Hunter E (May 2004). "An early stage of Mason-Pfizer monkey virus budding is regulated by the hydrophobicity of the Gag matrix domain core". J. Virol. 78 (10): 5023–31. doi:10.1128/jvi.78.10.5023-5031.2004. PMC 400380. PMID 15113883.
- ↑ Poon DT, Li G, Aldovini A (March 1998). "Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging". J. Virol. 72 (3): 1983–93. doi:10.1128/JVI.72.3.1983-1993.1998. PMC 109491. PMID 9499052.