| U3 | |
|---|---|
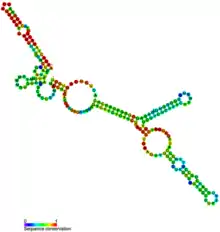 Metazoan U3 RNA secondary structure and sequence conservation | |
| Identifiers | |
| Symbol | U3 |
| Alt. Symbols | RNU3P2, |
| Rfam | RF00012 |
| NCBI Gene | 26844 |
| HGNC | 10176 |
| OMIM | 180710 |
| Other data | |
| RNA type | snoRNA |
| Domain(s) | Eukaryota; |
| PDB structures | PDBe |
In molecular biology, U3 snoRNA is a non-coding RNA found predominantly in the nucleolus. U3 has C/D box motifs that technically make it a member of the box C/D class of snoRNAs; however, unlike other C/D box snoRNAs, it has not been shown to direct 2'-O-methylation of other RNAs. Rather, U3 is thought to guide site-specific cleavage of ribosomal RNA (rRNA) during pre-rRNA processing.[1]
The box C/D element is a subset of the six short sequence elements found in all U3 snoRNAs, namely boxes A, A', B, C, C', and D.[2] The U3 snoRNA secondary structure is characterized by a small 5' domain (with boxes A and A'), and a larger 3' domain (with boxes B, C, C', and D), the two domains being linked by a single-stranded hinge. Boxes B and C form the B/C motif, which appears to be exclusive to U3 snoRNAs, and boxes C' and D form the C'/D motif. The latter is functionally similar to the C/D motifs found in other snoRNAs. The 5' domain and the hinge region act as a pre-rRNA-binding domain. The 3' domain has conserved protein-binding sites. Both the box B/C and box C'/D motifs are sufficient for nuclear retention of U3 snoRNA. The box C'/D motif is also necessary for nucleolar localization, stability and hyper-methylation of U3 snoRNA.[3] Both box B/C and C'/D motifs are involved in specific protein interactions and are necessary for the rRNA processing functions of U3 snoRNA.
Species-specific secondary structure models
S. cerevisiae secondary structure determined by chemical mapping of U3A RNA in a purified snoRNP is available.[4] A human structure model has also been proposed.[5] Like yeast and human, protozoan protist Entamoeba histolytica : a primitive eukaryote adopted the same conserved secondary structure of U3 snoRNA.[6] Four consensus structures specific to metazoa, fungi, plants and basal eukaryotes have been proposed.[7]
See also
References
- ↑ Cléry, A.; Senty-Ségault, V.; Leclerc, F.; Raué, A.; Branlant, C. (Feb 2007). "Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair". Molecular and Cellular Biology. 27 (4): 1191–1206. doi:10.1128/MCB.01287-06. ISSN 0270-7306. PMC 1800722. PMID 17145781.
- ↑ Zwieb, C (1997). "The uRNA database". Nucleic Acids Res. 25 (1): 102–103. doi:10.1093/nar/25.1.102. PMC 146409. PMID 9016512.
- ↑ Speckmann, W; Narayanan A; Terns R; Terns MP (1999). "Nuclear retention elements of U3 small nucleolar RNA". Mol Cell Biol. 19 (12): 8412–8421. doi:10.1128/MCB.19.12.8412. PMC 84939. PMID 10567566.
- ↑ Méreau A, Fournier R, Grégoire A, et al. (October 1997). "An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA". J. Mol. Biol. 273 (3): 552–71. doi:10.1006/jmbi.1997.1320. PMID 9356246.
- ↑ Granneman S, Vogelzangs J, Lührmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ (October 2004). "Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP". Mol. Cell. Biol. 24 (19): 8600–10. doi:10.1128/MCB.24.19.8600-8610.2004. PMC 516741. PMID 15367679.
- ↑ Srivastava A, Ahamad J, Ray AK, Kaur D, Bhattacharya A, Bhattacharya S (2014). Analysis of U3 snoRNA and small subunit processome components in the parasitic protist Entamoeba histolytica. Mol Biochem Parasitol. 193(2):82-92. doi: 10.1016/j.molbiopara.2014.03.001. Epub 2014 Mar 12
- ↑ Marz M, Stadler PF (2009). "Comparative analysis of eukaryotic U3 snoRNA". RNA Biol. 6 (5): 503–7. CiteSeerX 10.1.1.380.4189. doi:10.4161/rna.6.5.9607. PMID 19875933. S2CID 13055120.
External links
- Page for Small nucleolar RNA U3 at Rfam
- uRNADB: U3 page (archive)
- The UMASS snoRNAdb entry for U3
- The SGD entry for U3a
- The human snoRNAbase entry for U3